
Novosphingobium umbonatum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
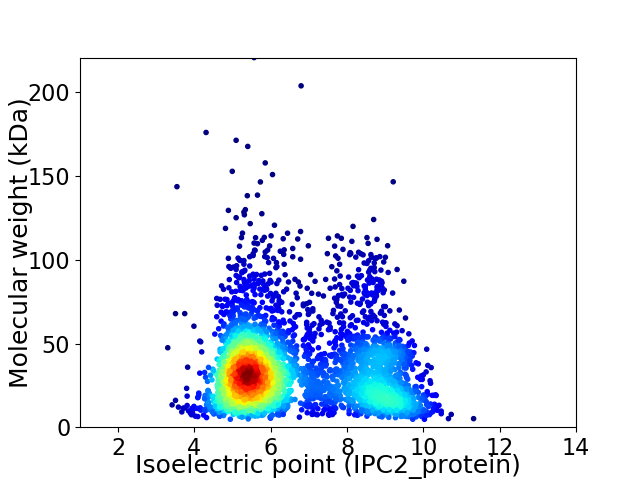
Virtual 2D-PAGE plot for 3564 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A437MXG9|A0A437MXG9_9SPHN Phosphatidylserine decarboxylase proenzyme OS=Novosphingobium umbonatum OX=1908524 GN=psd PE=3 SV=1
MM1 pKa = 7.32ATPIFVNEE9 pKa = 3.83LHH11 pKa = 6.55YY12 pKa = 11.49DD13 pKa = 3.49NAGTDD18 pKa = 3.08TGEE21 pKa = 4.68AIEE24 pKa = 4.27IAAPAGTNLAGWSIVLYY41 pKa = 10.9NGGTTGTAAVTYY53 pKa = 7.82GTIALSGTVANQGNGYY69 pKa = 8.0GTLSFAATGIQNGPVDD85 pKa = 4.53GFALVNASGTVVQFLSYY102 pKa = 10.33EE103 pKa = 4.28GVITASNGPASGMTSTDD120 pKa = 2.51IGVSQTGSQASGITLGLTGTGTTYY144 pKa = 11.36EE145 pKa = 4.04NFTWSTLTTGNFGSINNGQTFGSVAVPKK173 pKa = 10.72YY174 pKa = 10.85GEE176 pKa = 4.04LSIANASVTEE186 pKa = 4.43GNSGTQSLSFTVTRR200 pKa = 11.84ANGSDD205 pKa = 3.56GAVSASYY212 pKa = 9.62TVTLNSADD220 pKa = 4.0ASDD223 pKa = 4.23LAAGTTLTGTVSFAAGQTSANIVLGVLGDD252 pKa = 3.82TLVEE256 pKa = 4.21GDD258 pKa = 3.52EE259 pKa = 4.44TLTVTLSAPTGGATLGSVTTATGTITNDD287 pKa = 3.13DD288 pKa = 3.71TALPQAAAIAFINEE302 pKa = 3.39FHH304 pKa = 7.02YY305 pKa = 11.22DD306 pKa = 4.25DD307 pKa = 4.87IGTDD311 pKa = 2.98TGEE314 pKa = 4.54AIEE317 pKa = 4.26IAGTAGLNLAGWTLVLYY334 pKa = 10.36NGNGGAAYY342 pKa = 7.99KK343 pKa = 9.95TIALSGVIADD353 pKa = 3.76QDD355 pKa = 4.02SGFGTLSFAATDD367 pKa = 3.83IQNGSPDD374 pKa = 4.17GIALVDD380 pKa = 3.58ASGTVRR386 pKa = 11.84EE387 pKa = 4.26YY388 pKa = 10.6ISYY391 pKa = 9.48EE392 pKa = 3.88GVMTATSGAANGMVSEE408 pKa = 4.44DD409 pKa = 3.28VGVQEE414 pKa = 4.5NSAPEE419 pKa = 4.01GTSIQRR425 pKa = 11.84VGTGSDD431 pKa = 3.16PAAFTWVTGATSSFGSVNGSQDD453 pKa = 3.39FTPANPNGTLRR464 pKa = 11.84MGNASVVEE472 pKa = 4.24GDD474 pKa = 3.69SGTTAITFTVTRR486 pKa = 11.84SGGVTGAVSADD497 pKa = 3.44YY498 pKa = 11.37NVMFGTATADD508 pKa = 3.76DD509 pKa = 4.48LSGATSGTVNFATGQTSATITLHH532 pKa = 5.51VAADD536 pKa = 3.84TTGEE540 pKa = 3.95PDD542 pKa = 3.2EE543 pKa = 4.55MFSLVLSNAMGGADD557 pKa = 3.31IGRR560 pKa = 11.84SSGTGIIEE568 pKa = 4.33NDD570 pKa = 3.35DD571 pKa = 3.6KK572 pKa = 11.63TRR574 pKa = 11.84VTIMEE579 pKa = 4.11IQGEE583 pKa = 4.61SHH585 pKa = 6.17SSAYY589 pKa = 10.18AGQEE593 pKa = 3.89VTTKK597 pKa = 11.05GIVTAVDD604 pKa = 3.41GAGFYY609 pKa = 10.93LQDD612 pKa = 3.64AAGDD616 pKa = 4.22GNSRR620 pKa = 11.84TSDD623 pKa = 2.92AVFVYY628 pKa = 9.84TGTAPTVKK636 pKa = 9.72IGDD639 pKa = 4.58AIEE642 pKa = 4.07VTATVTEE649 pKa = 4.61YY650 pKa = 11.19VSSTTGSLSVTEE662 pKa = 4.01MTAPVITVEE671 pKa = 4.19SSGNALPAAVLIGPDD686 pKa = 3.6GLHH689 pKa = 6.54IPTANLEE696 pKa = 4.35DD697 pKa = 4.66DD698 pKa = 4.56GFTSFDD704 pKa = 3.97PATDD708 pKa = 4.16GLDD711 pKa = 3.25FWEE714 pKa = 4.61SLEE717 pKa = 3.84GMRR720 pKa = 11.84VEE722 pKa = 4.19IQNPVAIAPTATTYY736 pKa = 11.52GEE738 pKa = 4.42IYY740 pKa = 10.08TVASDD745 pKa = 4.3GAGHH749 pKa = 6.73TSSSGLSAEE758 pKa = 4.07STLAINGSYY767 pKa = 8.93STTGTGTNNSVNGDD781 pKa = 3.79YY782 pKa = 10.96NPEE785 pKa = 4.51KK786 pKa = 10.45IQIDD790 pKa = 3.62ADD792 pKa = 3.85VEE794 pKa = 4.54LGSGTAPLVNPGDD807 pKa = 3.74VLASVVGVVNYY818 pKa = 10.48AGGMYY823 pKa = 10.07EE824 pKa = 4.21VLATQAITIAQDD836 pKa = 3.21TTVAAQTTSLIAGTGQLAVANYY858 pKa = 10.01NVLNLDD864 pKa = 4.09PNDD867 pKa = 4.06ADD869 pKa = 4.01GDD871 pKa = 3.96TDD873 pKa = 3.78VASGRR878 pKa = 11.84FTMIAQDD885 pKa = 2.98IAFALGAPGIVVLEE899 pKa = 4.53EE900 pKa = 3.85IQDD903 pKa = 3.75NDD905 pKa = 3.17GSANTTTVSASLTLQMLVDD924 pKa = 4.95EE925 pKa = 5.65IYY927 pKa = 10.53AASGIQYY934 pKa = 10.8AWIDD938 pKa = 3.37NTYY941 pKa = 8.79ITDD944 pKa = 3.99DD945 pKa = 3.91NNGGEE950 pKa = 4.19PGGNIRR956 pKa = 11.84VAMIYY961 pKa = 10.24RR962 pKa = 11.84VDD964 pKa = 3.61TVDD967 pKa = 3.53LVEE970 pKa = 5.45GSVQTILPTSGSNPFDD986 pKa = 3.98GARR989 pKa = 11.84LPLVASFTFEE999 pKa = 4.28GEE1001 pKa = 3.98EE1002 pKa = 3.79LTVIGNHH1009 pKa = 5.3LTSKK1013 pKa = 10.7GGSTPLQGDD1022 pKa = 3.81VQPSTNAGEE1031 pKa = 4.34AARR1034 pKa = 11.84VAQADD1039 pKa = 3.83VVNDD1043 pKa = 3.74YY1044 pKa = 11.25VDD1046 pKa = 3.95SLLASDD1052 pKa = 4.88PSAHH1056 pKa = 6.57ILVSGDD1062 pKa = 3.28FNDD1065 pKa = 4.4FQFEE1069 pKa = 4.22EE1070 pKa = 4.37PLGVISGEE1078 pKa = 4.0LTLTDD1083 pKa = 3.83GVLTATDD1090 pKa = 3.91GVLTNLNTTIAAQDD1104 pKa = 3.31RR1105 pKa = 11.84YY1106 pKa = 11.36SYY1108 pKa = 10.62IFEE1111 pKa = 4.48GNAQEE1116 pKa = 4.22IDD1118 pKa = 4.05HH1119 pKa = 6.75ILVSDD1124 pKa = 3.99SLLDD1128 pKa = 3.76GAALDD1133 pKa = 3.68IVHH1136 pKa = 6.55VNTLTDD1142 pKa = 3.63RR1143 pKa = 11.84AASDD1147 pKa = 3.79HH1148 pKa = 7.01DD1149 pKa = 4.11PVVALLNLTTSLAVQTGTSGNDD1171 pKa = 3.31TLKK1174 pKa = 10.44GTDD1177 pKa = 3.21GRR1179 pKa = 11.84DD1180 pKa = 3.1RR1181 pKa = 11.84LIGGEE1186 pKa = 4.19GNDD1189 pKa = 3.69VLKK1192 pKa = 11.02GGAGADD1198 pKa = 3.3IMVGGAGDD1206 pKa = 3.48DD1207 pKa = 3.41AYY1209 pKa = 11.47YY1210 pKa = 10.85VDD1212 pKa = 3.92NAADD1216 pKa = 3.48IVTEE1220 pKa = 3.96AAGEE1224 pKa = 4.31GKK1226 pKa = 10.26DD1227 pKa = 3.17RR1228 pKa = 11.84VYY1230 pKa = 11.55ASVSYY1235 pKa = 9.51TLADD1239 pKa = 3.5TLEE1242 pKa = 4.26TLNLTGSDD1250 pKa = 3.38NLTGTGNAQDD1260 pKa = 3.82NQITGNDD1267 pKa = 4.14GDD1269 pKa = 3.91NLLYY1273 pKa = 11.03GLGGKK1278 pKa = 7.84DD1279 pKa = 3.42TLSGGAGADD1288 pKa = 3.32TLYY1291 pKa = 11.06GGEE1294 pKa = 4.73GNDD1297 pKa = 3.77TLKK1300 pKa = 11.43GEE1302 pKa = 4.62AGNDD1306 pKa = 3.73TLVGGAGADD1315 pKa = 4.02SLWGGAGADD1324 pKa = 2.89RR1325 pKa = 11.84FVFDD1329 pKa = 3.94VLEE1332 pKa = 4.32TATSKK1337 pKa = 11.09DD1338 pKa = 3.41AIGDD1342 pKa = 3.81FTSGEE1347 pKa = 4.14DD1348 pKa = 3.48KK1349 pKa = 11.15LVFDD1353 pKa = 5.23LSVFSSLSATGGVIDD1368 pKa = 4.99ASNIAFGGYY1377 pKa = 7.86ATTADD1382 pKa = 3.58QNLLYY1387 pKa = 10.22VASSGTLYY1395 pKa = 10.95YY1396 pKa = 10.32DD1397 pKa = 3.23TDD1399 pKa = 4.08GVGGADD1405 pKa = 2.61RR1406 pKa = 11.84VLIAVLTSKK1415 pKa = 10.42PVLSAADD1422 pKa = 3.83FVAAA1426 pKa = 4.98
MM1 pKa = 7.32ATPIFVNEE9 pKa = 3.83LHH11 pKa = 6.55YY12 pKa = 11.49DD13 pKa = 3.49NAGTDD18 pKa = 3.08TGEE21 pKa = 4.68AIEE24 pKa = 4.27IAAPAGTNLAGWSIVLYY41 pKa = 10.9NGGTTGTAAVTYY53 pKa = 7.82GTIALSGTVANQGNGYY69 pKa = 8.0GTLSFAATGIQNGPVDD85 pKa = 4.53GFALVNASGTVVQFLSYY102 pKa = 10.33EE103 pKa = 4.28GVITASNGPASGMTSTDD120 pKa = 2.51IGVSQTGSQASGITLGLTGTGTTYY144 pKa = 11.36EE145 pKa = 4.04NFTWSTLTTGNFGSINNGQTFGSVAVPKK173 pKa = 10.72YY174 pKa = 10.85GEE176 pKa = 4.04LSIANASVTEE186 pKa = 4.43GNSGTQSLSFTVTRR200 pKa = 11.84ANGSDD205 pKa = 3.56GAVSASYY212 pKa = 9.62TVTLNSADD220 pKa = 4.0ASDD223 pKa = 4.23LAAGTTLTGTVSFAAGQTSANIVLGVLGDD252 pKa = 3.82TLVEE256 pKa = 4.21GDD258 pKa = 3.52EE259 pKa = 4.44TLTVTLSAPTGGATLGSVTTATGTITNDD287 pKa = 3.13DD288 pKa = 3.71TALPQAAAIAFINEE302 pKa = 3.39FHH304 pKa = 7.02YY305 pKa = 11.22DD306 pKa = 4.25DD307 pKa = 4.87IGTDD311 pKa = 2.98TGEE314 pKa = 4.54AIEE317 pKa = 4.26IAGTAGLNLAGWTLVLYY334 pKa = 10.36NGNGGAAYY342 pKa = 7.99KK343 pKa = 9.95TIALSGVIADD353 pKa = 3.76QDD355 pKa = 4.02SGFGTLSFAATDD367 pKa = 3.83IQNGSPDD374 pKa = 4.17GIALVDD380 pKa = 3.58ASGTVRR386 pKa = 11.84EE387 pKa = 4.26YY388 pKa = 10.6ISYY391 pKa = 9.48EE392 pKa = 3.88GVMTATSGAANGMVSEE408 pKa = 4.44DD409 pKa = 3.28VGVQEE414 pKa = 4.5NSAPEE419 pKa = 4.01GTSIQRR425 pKa = 11.84VGTGSDD431 pKa = 3.16PAAFTWVTGATSSFGSVNGSQDD453 pKa = 3.39FTPANPNGTLRR464 pKa = 11.84MGNASVVEE472 pKa = 4.24GDD474 pKa = 3.69SGTTAITFTVTRR486 pKa = 11.84SGGVTGAVSADD497 pKa = 3.44YY498 pKa = 11.37NVMFGTATADD508 pKa = 3.76DD509 pKa = 4.48LSGATSGTVNFATGQTSATITLHH532 pKa = 5.51VAADD536 pKa = 3.84TTGEE540 pKa = 3.95PDD542 pKa = 3.2EE543 pKa = 4.55MFSLVLSNAMGGADD557 pKa = 3.31IGRR560 pKa = 11.84SSGTGIIEE568 pKa = 4.33NDD570 pKa = 3.35DD571 pKa = 3.6KK572 pKa = 11.63TRR574 pKa = 11.84VTIMEE579 pKa = 4.11IQGEE583 pKa = 4.61SHH585 pKa = 6.17SSAYY589 pKa = 10.18AGQEE593 pKa = 3.89VTTKK597 pKa = 11.05GIVTAVDD604 pKa = 3.41GAGFYY609 pKa = 10.93LQDD612 pKa = 3.64AAGDD616 pKa = 4.22GNSRR620 pKa = 11.84TSDD623 pKa = 2.92AVFVYY628 pKa = 9.84TGTAPTVKK636 pKa = 9.72IGDD639 pKa = 4.58AIEE642 pKa = 4.07VTATVTEE649 pKa = 4.61YY650 pKa = 11.19VSSTTGSLSVTEE662 pKa = 4.01MTAPVITVEE671 pKa = 4.19SSGNALPAAVLIGPDD686 pKa = 3.6GLHH689 pKa = 6.54IPTANLEE696 pKa = 4.35DD697 pKa = 4.66DD698 pKa = 4.56GFTSFDD704 pKa = 3.97PATDD708 pKa = 4.16GLDD711 pKa = 3.25FWEE714 pKa = 4.61SLEE717 pKa = 3.84GMRR720 pKa = 11.84VEE722 pKa = 4.19IQNPVAIAPTATTYY736 pKa = 11.52GEE738 pKa = 4.42IYY740 pKa = 10.08TVASDD745 pKa = 4.3GAGHH749 pKa = 6.73TSSSGLSAEE758 pKa = 4.07STLAINGSYY767 pKa = 8.93STTGTGTNNSVNGDD781 pKa = 3.79YY782 pKa = 10.96NPEE785 pKa = 4.51KK786 pKa = 10.45IQIDD790 pKa = 3.62ADD792 pKa = 3.85VEE794 pKa = 4.54LGSGTAPLVNPGDD807 pKa = 3.74VLASVVGVVNYY818 pKa = 10.48AGGMYY823 pKa = 10.07EE824 pKa = 4.21VLATQAITIAQDD836 pKa = 3.21TTVAAQTTSLIAGTGQLAVANYY858 pKa = 10.01NVLNLDD864 pKa = 4.09PNDD867 pKa = 4.06ADD869 pKa = 4.01GDD871 pKa = 3.96TDD873 pKa = 3.78VASGRR878 pKa = 11.84FTMIAQDD885 pKa = 2.98IAFALGAPGIVVLEE899 pKa = 4.53EE900 pKa = 3.85IQDD903 pKa = 3.75NDD905 pKa = 3.17GSANTTTVSASLTLQMLVDD924 pKa = 4.95EE925 pKa = 5.65IYY927 pKa = 10.53AASGIQYY934 pKa = 10.8AWIDD938 pKa = 3.37NTYY941 pKa = 8.79ITDD944 pKa = 3.99DD945 pKa = 3.91NNGGEE950 pKa = 4.19PGGNIRR956 pKa = 11.84VAMIYY961 pKa = 10.24RR962 pKa = 11.84VDD964 pKa = 3.61TVDD967 pKa = 3.53LVEE970 pKa = 5.45GSVQTILPTSGSNPFDD986 pKa = 3.98GARR989 pKa = 11.84LPLVASFTFEE999 pKa = 4.28GEE1001 pKa = 3.98EE1002 pKa = 3.79LTVIGNHH1009 pKa = 5.3LTSKK1013 pKa = 10.7GGSTPLQGDD1022 pKa = 3.81VQPSTNAGEE1031 pKa = 4.34AARR1034 pKa = 11.84VAQADD1039 pKa = 3.83VVNDD1043 pKa = 3.74YY1044 pKa = 11.25VDD1046 pKa = 3.95SLLASDD1052 pKa = 4.88PSAHH1056 pKa = 6.57ILVSGDD1062 pKa = 3.28FNDD1065 pKa = 4.4FQFEE1069 pKa = 4.22EE1070 pKa = 4.37PLGVISGEE1078 pKa = 4.0LTLTDD1083 pKa = 3.83GVLTATDD1090 pKa = 3.91GVLTNLNTTIAAQDD1104 pKa = 3.31RR1105 pKa = 11.84YY1106 pKa = 11.36SYY1108 pKa = 10.62IFEE1111 pKa = 4.48GNAQEE1116 pKa = 4.22IDD1118 pKa = 4.05HH1119 pKa = 6.75ILVSDD1124 pKa = 3.99SLLDD1128 pKa = 3.76GAALDD1133 pKa = 3.68IVHH1136 pKa = 6.55VNTLTDD1142 pKa = 3.63RR1143 pKa = 11.84AASDD1147 pKa = 3.79HH1148 pKa = 7.01DD1149 pKa = 4.11PVVALLNLTTSLAVQTGTSGNDD1171 pKa = 3.31TLKK1174 pKa = 10.44GTDD1177 pKa = 3.21GRR1179 pKa = 11.84DD1180 pKa = 3.1RR1181 pKa = 11.84LIGGEE1186 pKa = 4.19GNDD1189 pKa = 3.69VLKK1192 pKa = 11.02GGAGADD1198 pKa = 3.3IMVGGAGDD1206 pKa = 3.48DD1207 pKa = 3.41AYY1209 pKa = 11.47YY1210 pKa = 10.85VDD1212 pKa = 3.92NAADD1216 pKa = 3.48IVTEE1220 pKa = 3.96AAGEE1224 pKa = 4.31GKK1226 pKa = 10.26DD1227 pKa = 3.17RR1228 pKa = 11.84VYY1230 pKa = 11.55ASVSYY1235 pKa = 9.51TLADD1239 pKa = 3.5TLEE1242 pKa = 4.26TLNLTGSDD1250 pKa = 3.38NLTGTGNAQDD1260 pKa = 3.82NQITGNDD1267 pKa = 4.14GDD1269 pKa = 3.91NLLYY1273 pKa = 11.03GLGGKK1278 pKa = 7.84DD1279 pKa = 3.42TLSGGAGADD1288 pKa = 3.32TLYY1291 pKa = 11.06GGEE1294 pKa = 4.73GNDD1297 pKa = 3.77TLKK1300 pKa = 11.43GEE1302 pKa = 4.62AGNDD1306 pKa = 3.73TLVGGAGADD1315 pKa = 4.02SLWGGAGADD1324 pKa = 2.89RR1325 pKa = 11.84FVFDD1329 pKa = 3.94VLEE1332 pKa = 4.32TATSKK1337 pKa = 11.09DD1338 pKa = 3.41AIGDD1342 pKa = 3.81FTSGEE1347 pKa = 4.14DD1348 pKa = 3.48KK1349 pKa = 11.15LVFDD1353 pKa = 5.23LSVFSSLSATGGVIDD1368 pKa = 4.99ASNIAFGGYY1377 pKa = 7.86ATTADD1382 pKa = 3.58QNLLYY1387 pKa = 10.22VASSGTLYY1395 pKa = 10.95YY1396 pKa = 10.32DD1397 pKa = 3.23TDD1399 pKa = 4.08GVGGADD1405 pKa = 2.61RR1406 pKa = 11.84VLIAVLTSKK1415 pKa = 10.42PVLSAADD1422 pKa = 3.83FVAAA1426 pKa = 4.98
Molecular weight: 143.69 kDa
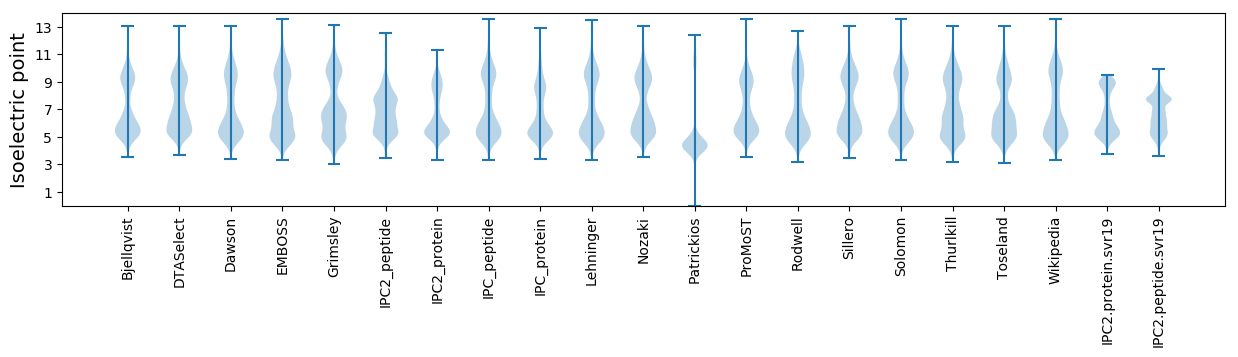
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S2YAN4|A0A3S2YAN4_9SPHN Sporulation protein SsgA OS=Novosphingobium umbonatum OX=1908524 GN=EOE18_05325 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 8.17VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.79NLSAA44 pKa = 4.73
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 8.17VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.79NLSAA44 pKa = 4.73
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1221789 |
41 |
1997 |
342.8 |
36.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.422 ± 0.053 | 0.839 ± 0.014 |
5.48 ± 0.03 | 5.141 ± 0.036 |
3.481 ± 0.022 | 8.87 ± 0.043 |
2.107 ± 0.019 | 4.693 ± 0.026 |
3.363 ± 0.027 | 10.131 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.603 ± 0.02 | 2.795 ± 0.031 |
5.202 ± 0.03 | 3.69 ± 0.025 |
6.555 ± 0.041 | 5.491 ± 0.035 |
5.143 ± 0.037 | 7.099 ± 0.032 |
1.511 ± 0.019 | 2.383 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
