
Psychromicrobium lacuslunae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Psychromicrobium
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
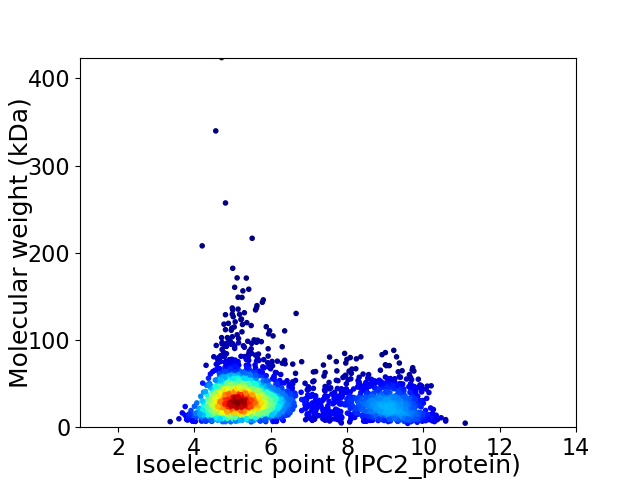
Virtual 2D-PAGE plot for 2887 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
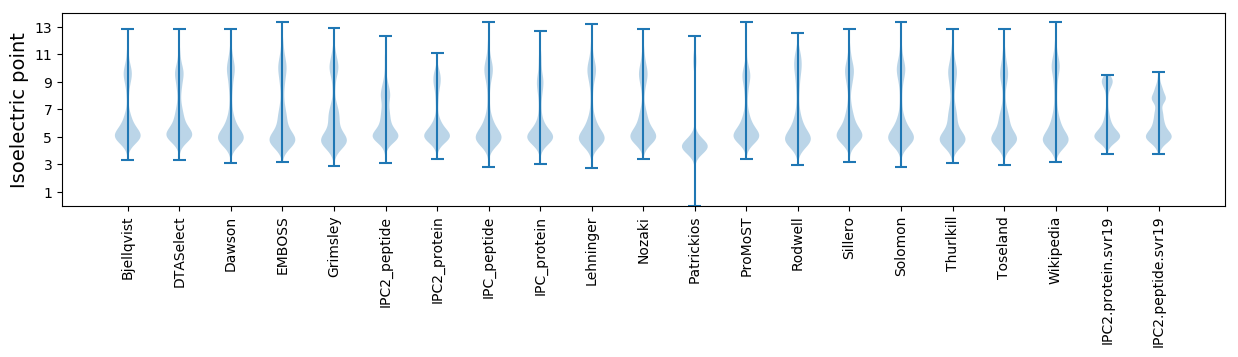
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D4BYY9|A0A0D4BYY9_9MICC Uncharacterized protein OS=Psychromicrobium lacuslunae OX=1618207 GN=UM93_07055 PE=4 SV=1
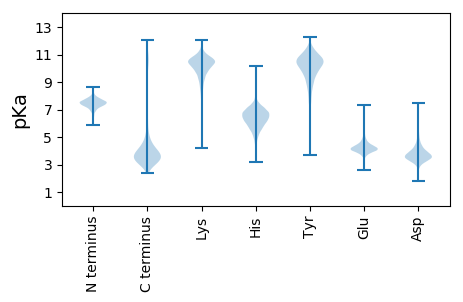
MM1 pKa = 7.68PKK3 pKa = 10.56ALIIVDD9 pKa = 3.88VQNDD13 pKa = 3.63FCEE16 pKa = 4.91GGSLAVSGGEE26 pKa = 3.96EE27 pKa = 4.08VATGISEE34 pKa = 4.84LLDD37 pKa = 3.34STFGAEE43 pKa = 4.06SRR45 pKa = 11.84EE46 pKa = 3.72FDD48 pKa = 4.02YY49 pKa = 11.64VLATQDD55 pKa = 2.96WHH57 pKa = 6.99IDD59 pKa = 3.55PGSHH63 pKa = 7.03FSEE66 pKa = 4.47TPDD69 pKa = 5.8FIDD72 pKa = 3.23SWPVHH77 pKa = 5.93CVAGSKK83 pKa = 10.15GAQLHH88 pKa = 6.53PNLDD92 pKa = 3.59TEE94 pKa = 4.48YY95 pKa = 10.48VDD97 pKa = 4.28AYY99 pKa = 10.22FRR101 pKa = 11.84KK102 pKa = 10.09GKK104 pKa = 9.54YY105 pKa = 7.67QAAYY109 pKa = 10.34SGFEE113 pKa = 4.02GFLAPEE119 pKa = 4.68DD120 pKa = 4.12EE121 pKa = 4.87VPTGPRR127 pKa = 11.84NPEE130 pKa = 3.51QTEE133 pKa = 3.89NPEE136 pKa = 4.03NSEE139 pKa = 4.02SSGDD143 pKa = 3.54VGYY146 pKa = 10.94DD147 pKa = 3.4GDD149 pKa = 5.37DD150 pKa = 4.48DD151 pKa = 5.5GPSLDD156 pKa = 3.46EE157 pKa = 3.92WLQEE161 pKa = 4.07HH162 pKa = 6.29EE163 pKa = 4.38VDD165 pKa = 3.68SVSIVGLATDD175 pKa = 3.58YY176 pKa = 10.89CVRR179 pKa = 11.84ATALDD184 pKa = 3.82AVSAGYY190 pKa = 10.01EE191 pKa = 4.02VTLLSEE197 pKa = 4.56LCAGIDD203 pKa = 3.64PAGSQAALTEE213 pKa = 4.12LTEE216 pKa = 4.59AGVEE220 pKa = 4.36III222 pKa = 5.11
MM1 pKa = 7.68PKK3 pKa = 10.56ALIIVDD9 pKa = 3.88VQNDD13 pKa = 3.63FCEE16 pKa = 4.91GGSLAVSGGEE26 pKa = 3.96EE27 pKa = 4.08VATGISEE34 pKa = 4.84LLDD37 pKa = 3.34STFGAEE43 pKa = 4.06SRR45 pKa = 11.84EE46 pKa = 3.72FDD48 pKa = 4.02YY49 pKa = 11.64VLATQDD55 pKa = 2.96WHH57 pKa = 6.99IDD59 pKa = 3.55PGSHH63 pKa = 7.03FSEE66 pKa = 4.47TPDD69 pKa = 5.8FIDD72 pKa = 3.23SWPVHH77 pKa = 5.93CVAGSKK83 pKa = 10.15GAQLHH88 pKa = 6.53PNLDD92 pKa = 3.59TEE94 pKa = 4.48YY95 pKa = 10.48VDD97 pKa = 4.28AYY99 pKa = 10.22FRR101 pKa = 11.84KK102 pKa = 10.09GKK104 pKa = 9.54YY105 pKa = 7.67QAAYY109 pKa = 10.34SGFEE113 pKa = 4.02GFLAPEE119 pKa = 4.68DD120 pKa = 4.12EE121 pKa = 4.87VPTGPRR127 pKa = 11.84NPEE130 pKa = 3.51QTEE133 pKa = 3.89NPEE136 pKa = 4.03NSEE139 pKa = 4.02SSGDD143 pKa = 3.54VGYY146 pKa = 10.94DD147 pKa = 3.4GDD149 pKa = 5.37DD150 pKa = 4.48DD151 pKa = 5.5GPSLDD156 pKa = 3.46EE157 pKa = 3.92WLQEE161 pKa = 4.07HH162 pKa = 6.29EE163 pKa = 4.38VDD165 pKa = 3.68SVSIVGLATDD175 pKa = 3.58YY176 pKa = 10.89CVRR179 pKa = 11.84ATALDD184 pKa = 3.82AVSAGYY190 pKa = 10.01EE191 pKa = 4.02VTLLSEE197 pKa = 4.56LCAGIDD203 pKa = 3.64PAGSQAALTEE213 pKa = 4.12LTEE216 pKa = 4.59AGVEE220 pKa = 4.36III222 pKa = 5.11
Molecular weight: 23.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D4BZ54|A0A0D4BZ54_9MICC HTH tetR-type domain-containing protein OS=Psychromicrobium lacuslunae OX=1618207 GN=UM93_08905 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.31VRR4 pKa = 11.84NSLRR8 pKa = 11.84ALKK11 pKa = 9.71KK12 pKa = 10.76LPGAQIVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84TFVINKK30 pKa = 8.58NNPRR34 pKa = 11.84MKK36 pKa = 10.38ARR38 pKa = 11.84QGG40 pKa = 3.24
MM1 pKa = 7.61KK2 pKa = 10.31VRR4 pKa = 11.84NSLRR8 pKa = 11.84ALKK11 pKa = 9.71KK12 pKa = 10.76LPGAQIVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84TFVINKK30 pKa = 8.58NNPRR34 pKa = 11.84MKK36 pKa = 10.38ARR38 pKa = 11.84QGG40 pKa = 3.24
Molecular weight: 4.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
921680 |
37 |
3915 |
319.3 |
34.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.35 ± 0.062 | 0.577 ± 0.01 |
5.123 ± 0.033 | 5.977 ± 0.055 |
3.343 ± 0.032 | 8.411 ± 0.042 |
1.884 ± 0.022 | 4.807 ± 0.039 |
3.126 ± 0.041 | 10.957 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.832 ± 0.019 | 2.75 ± 0.035 |
5.03 ± 0.027 | 4.033 ± 0.03 |
6.065 ± 0.047 | 6.783 ± 0.045 |
5.422 ± 0.049 | 7.902 ± 0.04 |
1.464 ± 0.024 | 2.162 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
