
Streptomyces sp. CB01249
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
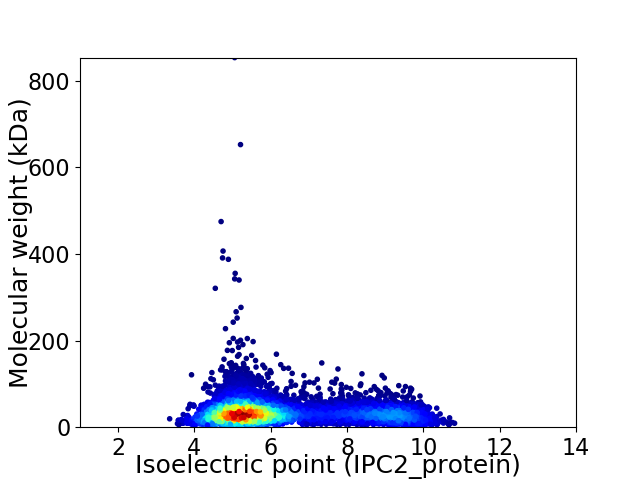
Virtual 2D-PAGE plot for 6535 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q5DY20|A0A1Q5DY20_9ACTN Sugar ABC transporter permease OS=Streptomyces sp. CB01249 OX=1703929 GN=AMK18_19150 PE=3 SV=1
MM1 pKa = 7.31SKK3 pKa = 10.48EE4 pKa = 3.79NPAEE8 pKa = 3.88QHH10 pKa = 5.98HH11 pKa = 6.75GGWQPTPQGGDD22 pKa = 3.21YY23 pKa = 10.94DD24 pKa = 4.31GEE26 pKa = 4.08ATAFVQLPPEE36 pKa = 4.49DD37 pKa = 4.12LADD40 pKa = 3.86IPLAAPGHH48 pKa = 6.64GYY50 pKa = 7.51TPPMILPLTPAADD63 pKa = 4.33RR64 pKa = 11.84DD65 pKa = 3.57PAAAGNWVVQTPEE78 pKa = 3.88QQPGQPAPDD87 pKa = 3.5AVHH90 pKa = 6.67WPDD93 pKa = 3.91PNQQSGYY100 pKa = 9.97GYY102 pKa = 8.45PHH104 pKa = 7.64PEE106 pKa = 4.1DD107 pKa = 3.57QQPTGSYY114 pKa = 9.58QEE116 pKa = 4.62PYY118 pKa = 9.08GTDD121 pKa = 3.4PAATGQWAVDD131 pKa = 3.74TSSWPAPAAPLSDD144 pKa = 4.64DD145 pKa = 3.67SGEE148 pKa = 4.74LYY150 pKa = 10.2TPPPVADD157 pKa = 4.22WYY159 pKa = 10.88ADD161 pKa = 3.98RR162 pKa = 11.84PATLPGGAPAPWAVPDD178 pKa = 3.8AAEE181 pKa = 4.1AAVPEE186 pKa = 4.33AAEE189 pKa = 4.18VTEE192 pKa = 4.6PEE194 pKa = 4.65AVDD197 pKa = 3.9APADD201 pKa = 3.76ADD203 pKa = 3.96EE204 pKa = 5.05PADD207 pKa = 3.9ALAGGPAEE215 pKa = 4.46SAPEE219 pKa = 3.8AAEE222 pKa = 4.1PDD224 pKa = 3.73AEE226 pKa = 4.36QPAPEE231 pKa = 4.76ADD233 pKa = 3.43ALAAADD239 pKa = 4.14PVAAEE244 pKa = 4.61PGAEE248 pKa = 3.96PAEE251 pKa = 4.33PAGEE255 pKa = 4.03AVPDD259 pKa = 3.86AEE261 pKa = 4.41VPEE264 pKa = 5.05AEE266 pKa = 3.96AVQPAEE272 pKa = 4.36VPEE275 pKa = 4.43APAPEE280 pKa = 4.9GPPLPSEE287 pKa = 4.7HH288 pKa = 6.88PAASYY293 pKa = 9.99VLHH296 pKa = 6.36VNGVDD301 pKa = 4.64RR302 pKa = 11.84PVTDD306 pKa = 2.96AWIGEE311 pKa = 4.15SLLYY315 pKa = 10.18VLRR318 pKa = 11.84EE319 pKa = 3.94RR320 pKa = 11.84LGLAGAKK327 pKa = 9.72DD328 pKa = 3.75GCSQGEE334 pKa = 4.12CGACNVQVDD343 pKa = 3.94GRR345 pKa = 11.84LVASCLVPAATTAGSEE361 pKa = 3.99VRR363 pKa = 11.84TVEE366 pKa = 3.9GLAVDD371 pKa = 4.82GEE373 pKa = 4.31PSDD376 pKa = 3.93VQRR379 pKa = 11.84ALADD383 pKa = 3.82CGAVQCGFCIPGMAMTVHH401 pKa = 7.14DD402 pKa = 5.1LLEE405 pKa = 4.95GNHH408 pKa = 6.22RR409 pKa = 11.84PSEE412 pKa = 4.25LEE414 pKa = 3.67TRR416 pKa = 11.84RR417 pKa = 11.84ALCGNLCRR425 pKa = 11.84CSGYY429 pKa = 10.39RR430 pKa = 11.84GVLDD434 pKa = 3.98AVNEE438 pKa = 4.35VIAGRR443 pKa = 11.84EE444 pKa = 3.72AAAEE448 pKa = 3.96AVAEE452 pKa = 4.28EE453 pKa = 4.48TGSGEE458 pKa = 4.07PEE460 pKa = 4.27EE461 pKa = 4.52PRR463 pKa = 11.84IPHH466 pKa = 5.36QAAPGAGSVQAHH478 pKa = 4.76QQDD481 pKa = 3.47GGMAA485 pKa = 3.59
MM1 pKa = 7.31SKK3 pKa = 10.48EE4 pKa = 3.79NPAEE8 pKa = 3.88QHH10 pKa = 5.98HH11 pKa = 6.75GGWQPTPQGGDD22 pKa = 3.21YY23 pKa = 10.94DD24 pKa = 4.31GEE26 pKa = 4.08ATAFVQLPPEE36 pKa = 4.49DD37 pKa = 4.12LADD40 pKa = 3.86IPLAAPGHH48 pKa = 6.64GYY50 pKa = 7.51TPPMILPLTPAADD63 pKa = 4.33RR64 pKa = 11.84DD65 pKa = 3.57PAAAGNWVVQTPEE78 pKa = 3.88QQPGQPAPDD87 pKa = 3.5AVHH90 pKa = 6.67WPDD93 pKa = 3.91PNQQSGYY100 pKa = 9.97GYY102 pKa = 8.45PHH104 pKa = 7.64PEE106 pKa = 4.1DD107 pKa = 3.57QQPTGSYY114 pKa = 9.58QEE116 pKa = 4.62PYY118 pKa = 9.08GTDD121 pKa = 3.4PAATGQWAVDD131 pKa = 3.74TSSWPAPAAPLSDD144 pKa = 4.64DD145 pKa = 3.67SGEE148 pKa = 4.74LYY150 pKa = 10.2TPPPVADD157 pKa = 4.22WYY159 pKa = 10.88ADD161 pKa = 3.98RR162 pKa = 11.84PATLPGGAPAPWAVPDD178 pKa = 3.8AAEE181 pKa = 4.1AAVPEE186 pKa = 4.33AAEE189 pKa = 4.18VTEE192 pKa = 4.6PEE194 pKa = 4.65AVDD197 pKa = 3.9APADD201 pKa = 3.76ADD203 pKa = 3.96EE204 pKa = 5.05PADD207 pKa = 3.9ALAGGPAEE215 pKa = 4.46SAPEE219 pKa = 3.8AAEE222 pKa = 4.1PDD224 pKa = 3.73AEE226 pKa = 4.36QPAPEE231 pKa = 4.76ADD233 pKa = 3.43ALAAADD239 pKa = 4.14PVAAEE244 pKa = 4.61PGAEE248 pKa = 3.96PAEE251 pKa = 4.33PAGEE255 pKa = 4.03AVPDD259 pKa = 3.86AEE261 pKa = 4.41VPEE264 pKa = 5.05AEE266 pKa = 3.96AVQPAEE272 pKa = 4.36VPEE275 pKa = 4.43APAPEE280 pKa = 4.9GPPLPSEE287 pKa = 4.7HH288 pKa = 6.88PAASYY293 pKa = 9.99VLHH296 pKa = 6.36VNGVDD301 pKa = 4.64RR302 pKa = 11.84PVTDD306 pKa = 2.96AWIGEE311 pKa = 4.15SLLYY315 pKa = 10.18VLRR318 pKa = 11.84EE319 pKa = 3.94RR320 pKa = 11.84LGLAGAKK327 pKa = 9.72DD328 pKa = 3.75GCSQGEE334 pKa = 4.12CGACNVQVDD343 pKa = 3.94GRR345 pKa = 11.84LVASCLVPAATTAGSEE361 pKa = 3.99VRR363 pKa = 11.84TVEE366 pKa = 3.9GLAVDD371 pKa = 4.82GEE373 pKa = 4.31PSDD376 pKa = 3.93VQRR379 pKa = 11.84ALADD383 pKa = 3.82CGAVQCGFCIPGMAMTVHH401 pKa = 7.14DD402 pKa = 5.1LLEE405 pKa = 4.95GNHH408 pKa = 6.22RR409 pKa = 11.84PSEE412 pKa = 4.25LEE414 pKa = 3.67TRR416 pKa = 11.84RR417 pKa = 11.84ALCGNLCRR425 pKa = 11.84CSGYY429 pKa = 10.39RR430 pKa = 11.84GVLDD434 pKa = 3.98AVNEE438 pKa = 4.35VIAGRR443 pKa = 11.84EE444 pKa = 3.72AAAEE448 pKa = 3.96AVAEE452 pKa = 4.28EE453 pKa = 4.48TGSGEE458 pKa = 4.07PEE460 pKa = 4.27EE461 pKa = 4.52PRR463 pKa = 11.84IPHH466 pKa = 5.36QAAPGAGSVQAHH478 pKa = 4.76QQDD481 pKa = 3.47GGMAA485 pKa = 3.59
Molecular weight: 49.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q5E2J8|A0A1Q5E2J8_9ACTN 3-methyladenine DNA glycosylase OS=Streptomyces sp. CB01249 OX=1703929 GN=AMK18_12655 PE=4 SV=1
MM1 pKa = 8.03ILEE4 pKa = 4.28TVSAVVLGVALSWAALRR21 pKa = 11.84SLPGRR26 pKa = 11.84LPARR30 pKa = 11.84RR31 pKa = 11.84TVFGTGALGGLFGASLMHH49 pKa = 6.54SVLGPHH55 pKa = 6.58HH56 pKa = 6.65AAAVVVGAVLIAAVTVSLLIRR77 pKa = 11.84PRR79 pKa = 11.84SRR81 pKa = 11.84RR82 pKa = 11.84LRR84 pKa = 11.84RR85 pKa = 11.84SAAAA89 pKa = 3.76
MM1 pKa = 8.03ILEE4 pKa = 4.28TVSAVVLGVALSWAALRR21 pKa = 11.84SLPGRR26 pKa = 11.84LPARR30 pKa = 11.84RR31 pKa = 11.84TVFGTGALGGLFGASLMHH49 pKa = 6.54SVLGPHH55 pKa = 6.58HH56 pKa = 6.65AAAVVVGAVLIAAVTVSLLIRR77 pKa = 11.84PRR79 pKa = 11.84SRR81 pKa = 11.84RR82 pKa = 11.84LRR84 pKa = 11.84RR85 pKa = 11.84SAAAA89 pKa = 3.76
Molecular weight: 9.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2242174 |
33 |
7970 |
343.1 |
36.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.807 ± 0.049 | 0.761 ± 0.009 |
6.09 ± 0.024 | 5.658 ± 0.028 |
2.739 ± 0.018 | 9.596 ± 0.03 |
2.238 ± 0.015 | 3.129 ± 0.022 |
2.219 ± 0.028 | 10.355 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.728 ± 0.012 | 1.801 ± 0.019 |
6.129 ± 0.026 | 2.658 ± 0.02 |
7.828 ± 0.034 | 5.009 ± 0.021 |
6.226 ± 0.027 | 8.396 ± 0.032 |
1.491 ± 0.013 | 2.14 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
