
Capybara microvirus Cap1_SP_60
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
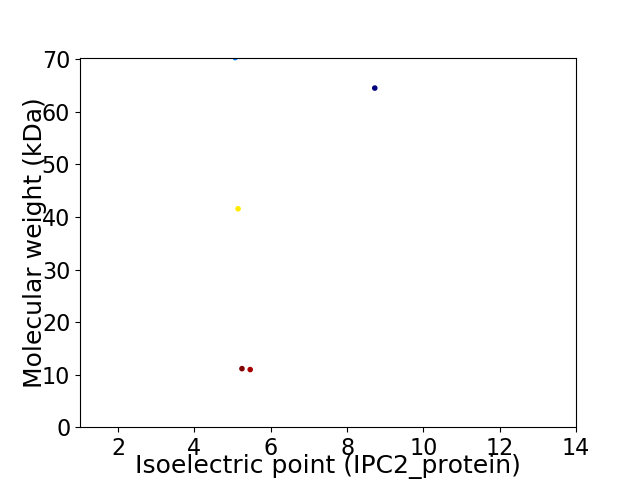
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
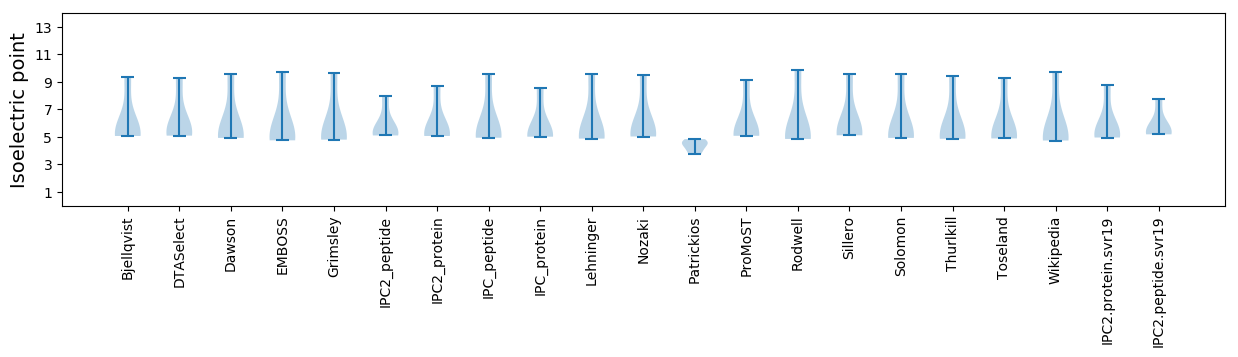
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W4N9|A0A4P8W4N9_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_60 OX=2584785 PE=4 SV=1
MM1 pKa = 7.53ATRR4 pKa = 11.84KK5 pKa = 9.31FQFEE9 pKa = 4.39TMTAAGKK16 pKa = 10.58KK17 pKa = 9.91SVFDD21 pKa = 3.31MSFNHH26 pKa = 7.18IGTQDD31 pKa = 3.41FGYY34 pKa = 10.45LKK36 pKa = 10.16PIGMNFLVPGDD47 pKa = 3.45EE48 pKa = 3.88WKK50 pKa = 10.81INITQFTRR58 pKa = 11.84LMPMPCPTFGKK69 pKa = 9.53VDD71 pKa = 3.19TRR73 pKa = 11.84IRR75 pKa = 11.84AFFVPFQTILNGWNDD90 pKa = 4.48FIANRR95 pKa = 11.84VSPSTGGEE103 pKa = 3.44IAIPHH108 pKa = 5.23VTNEE112 pKa = 3.51WFTNWFVKK120 pKa = 10.43KK121 pKa = 10.29NAFILEE127 pKa = 4.35DD128 pKa = 3.52QDD130 pKa = 4.27LPLYY134 pKa = 10.64QEE136 pKa = 4.92GSWDD140 pKa = 3.65YY141 pKa = 10.53YY142 pKa = 11.23DD143 pKa = 4.56IIVRR147 pKa = 11.84EE148 pKa = 4.09NDD150 pKa = 4.09GLWHH154 pKa = 6.49GLNLTPLGKK163 pKa = 9.79RR164 pKa = 11.84FYY166 pKa = 11.63DD167 pKa = 3.38MLLGCGINIQMCAYY181 pKa = 9.97KK182 pKa = 9.65STTEE186 pKa = 3.66EE187 pKa = 4.68AYY189 pKa = 9.81TEE191 pKa = 3.68IAQRR195 pKa = 11.84KK196 pKa = 8.79FSVLPIAGLYY206 pKa = 8.97RR207 pKa = 11.84WYY209 pKa = 9.61TDD211 pKa = 3.2WIVPSRR217 pKa = 11.84FVDD220 pKa = 3.62EE221 pKa = 4.18YY222 pKa = 10.65MPIRR226 pKa = 11.84MLFVANEE233 pKa = 3.87WSGEE237 pKa = 4.06EE238 pKa = 4.97LFTAMDD244 pKa = 3.33QIYY247 pKa = 10.09NVNPAALVLSTFYY260 pKa = 11.23ADD262 pKa = 6.02DD263 pKa = 3.74MFSCAFKK270 pKa = 11.27NPFGLEE276 pKa = 3.92NQSMYY281 pKa = 10.84DD282 pKa = 3.68YY283 pKa = 10.82NIQNPAIYY291 pKa = 9.87SSEE294 pKa = 3.94GTHH297 pKa = 5.6MLGAEE302 pKa = 5.33AITSIDD308 pKa = 3.59PTLQAAKK315 pKa = 9.89QGARR319 pKa = 11.84IKK321 pKa = 10.48RR322 pKa = 11.84GEE324 pKa = 4.25GEE326 pKa = 4.12PSNGSTWGINHH337 pKa = 5.91FTIKK341 pKa = 10.52SLGVLQDD348 pKa = 3.4LLNRR352 pKa = 11.84GKK354 pKa = 10.21IAGTKK359 pKa = 8.34IQDD362 pKa = 4.23YY363 pKa = 10.41LWEE366 pKa = 4.43TYY368 pKa = 10.21GIKK371 pKa = 10.68ANSDD375 pKa = 3.65EE376 pKa = 5.04LNLSLYY382 pKa = 10.38LGSSQDD388 pKa = 3.19IIKK391 pKa = 10.44IGDD394 pKa = 3.48VMATAGTEE402 pKa = 4.2VNEE405 pKa = 4.34LGQFAGKK412 pKa = 10.5GIGSMRR418 pKa = 11.84FGCTCKK424 pKa = 10.44SGNKK428 pKa = 9.55HH429 pKa = 5.51GLLYY433 pKa = 10.69LCSEE437 pKa = 4.52INVDD441 pKa = 3.1PSYY444 pKa = 11.26YY445 pKa = 9.94QGEE448 pKa = 4.37KK449 pKa = 10.48PEE451 pKa = 3.93MRR453 pKa = 11.84QLEE456 pKa = 4.26RR457 pKa = 11.84DD458 pKa = 3.49DD459 pKa = 5.31FYY461 pKa = 11.62QPEE464 pKa = 4.16FDD466 pKa = 3.77NLGVEE471 pKa = 5.38AISKK475 pKa = 9.74AQIKK479 pKa = 10.59AEE481 pKa = 3.79WDD483 pKa = 3.55NNGMNMYY490 pKa = 9.58GYY492 pKa = 9.64PIDD495 pKa = 4.33SPDD498 pKa = 3.86SIFGFTPRR506 pKa = 11.84YY507 pKa = 9.58SKK509 pKa = 11.46YY510 pKa = 10.67KK511 pKa = 9.97FNNDD515 pKa = 3.11IISGDD520 pKa = 3.39FRR522 pKa = 11.84LQSSRR527 pKa = 11.84TLRR530 pKa = 11.84SWYY533 pKa = 10.12LSRR536 pKa = 11.84NLEE539 pKa = 4.22YY540 pKa = 9.66ATKK543 pKa = 9.34GTWTNINSNFCRR555 pKa = 11.84QLVDD559 pKa = 4.06IEE561 pKa = 4.41INNYY565 pKa = 10.96DD566 pKa = 3.8YY567 pKa = 11.15MFNAANNNSDD577 pKa = 2.56HH578 pKa = 6.82FYY580 pKa = 10.99NVFSVGIKK588 pKa = 10.11ALRR591 pKa = 11.84PMEE594 pKa = 4.67TISKK598 pKa = 10.27ALEE601 pKa = 4.25TEE603 pKa = 4.13DD604 pKa = 3.74TNDD607 pKa = 2.91MGRR610 pKa = 11.84KK611 pKa = 9.22VRR613 pKa = 11.84VSMNN617 pKa = 3.02
MM1 pKa = 7.53ATRR4 pKa = 11.84KK5 pKa = 9.31FQFEE9 pKa = 4.39TMTAAGKK16 pKa = 10.58KK17 pKa = 9.91SVFDD21 pKa = 3.31MSFNHH26 pKa = 7.18IGTQDD31 pKa = 3.41FGYY34 pKa = 10.45LKK36 pKa = 10.16PIGMNFLVPGDD47 pKa = 3.45EE48 pKa = 3.88WKK50 pKa = 10.81INITQFTRR58 pKa = 11.84LMPMPCPTFGKK69 pKa = 9.53VDD71 pKa = 3.19TRR73 pKa = 11.84IRR75 pKa = 11.84AFFVPFQTILNGWNDD90 pKa = 4.48FIANRR95 pKa = 11.84VSPSTGGEE103 pKa = 3.44IAIPHH108 pKa = 5.23VTNEE112 pKa = 3.51WFTNWFVKK120 pKa = 10.43KK121 pKa = 10.29NAFILEE127 pKa = 4.35DD128 pKa = 3.52QDD130 pKa = 4.27LPLYY134 pKa = 10.64QEE136 pKa = 4.92GSWDD140 pKa = 3.65YY141 pKa = 10.53YY142 pKa = 11.23DD143 pKa = 4.56IIVRR147 pKa = 11.84EE148 pKa = 4.09NDD150 pKa = 4.09GLWHH154 pKa = 6.49GLNLTPLGKK163 pKa = 9.79RR164 pKa = 11.84FYY166 pKa = 11.63DD167 pKa = 3.38MLLGCGINIQMCAYY181 pKa = 9.97KK182 pKa = 9.65STTEE186 pKa = 3.66EE187 pKa = 4.68AYY189 pKa = 9.81TEE191 pKa = 3.68IAQRR195 pKa = 11.84KK196 pKa = 8.79FSVLPIAGLYY206 pKa = 8.97RR207 pKa = 11.84WYY209 pKa = 9.61TDD211 pKa = 3.2WIVPSRR217 pKa = 11.84FVDD220 pKa = 3.62EE221 pKa = 4.18YY222 pKa = 10.65MPIRR226 pKa = 11.84MLFVANEE233 pKa = 3.87WSGEE237 pKa = 4.06EE238 pKa = 4.97LFTAMDD244 pKa = 3.33QIYY247 pKa = 10.09NVNPAALVLSTFYY260 pKa = 11.23ADD262 pKa = 6.02DD263 pKa = 3.74MFSCAFKK270 pKa = 11.27NPFGLEE276 pKa = 3.92NQSMYY281 pKa = 10.84DD282 pKa = 3.68YY283 pKa = 10.82NIQNPAIYY291 pKa = 9.87SSEE294 pKa = 3.94GTHH297 pKa = 5.6MLGAEE302 pKa = 5.33AITSIDD308 pKa = 3.59PTLQAAKK315 pKa = 9.89QGARR319 pKa = 11.84IKK321 pKa = 10.48RR322 pKa = 11.84GEE324 pKa = 4.25GEE326 pKa = 4.12PSNGSTWGINHH337 pKa = 5.91FTIKK341 pKa = 10.52SLGVLQDD348 pKa = 3.4LLNRR352 pKa = 11.84GKK354 pKa = 10.21IAGTKK359 pKa = 8.34IQDD362 pKa = 4.23YY363 pKa = 10.41LWEE366 pKa = 4.43TYY368 pKa = 10.21GIKK371 pKa = 10.68ANSDD375 pKa = 3.65EE376 pKa = 5.04LNLSLYY382 pKa = 10.38LGSSQDD388 pKa = 3.19IIKK391 pKa = 10.44IGDD394 pKa = 3.48VMATAGTEE402 pKa = 4.2VNEE405 pKa = 4.34LGQFAGKK412 pKa = 10.5GIGSMRR418 pKa = 11.84FGCTCKK424 pKa = 10.44SGNKK428 pKa = 9.55HH429 pKa = 5.51GLLYY433 pKa = 10.69LCSEE437 pKa = 4.52INVDD441 pKa = 3.1PSYY444 pKa = 11.26YY445 pKa = 9.94QGEE448 pKa = 4.37KK449 pKa = 10.48PEE451 pKa = 3.93MRR453 pKa = 11.84QLEE456 pKa = 4.26RR457 pKa = 11.84DD458 pKa = 3.49DD459 pKa = 5.31FYY461 pKa = 11.62QPEE464 pKa = 4.16FDD466 pKa = 3.77NLGVEE471 pKa = 5.38AISKK475 pKa = 9.74AQIKK479 pKa = 10.59AEE481 pKa = 3.79WDD483 pKa = 3.55NNGMNMYY490 pKa = 9.58GYY492 pKa = 9.64PIDD495 pKa = 4.33SPDD498 pKa = 3.86SIFGFTPRR506 pKa = 11.84YY507 pKa = 9.58SKK509 pKa = 11.46YY510 pKa = 10.67KK511 pKa = 9.97FNNDD515 pKa = 3.11IISGDD520 pKa = 3.39FRR522 pKa = 11.84LQSSRR527 pKa = 11.84TLRR530 pKa = 11.84SWYY533 pKa = 10.12LSRR536 pKa = 11.84NLEE539 pKa = 4.22YY540 pKa = 9.66ATKK543 pKa = 9.34GTWTNINSNFCRR555 pKa = 11.84QLVDD559 pKa = 4.06IEE561 pKa = 4.41INNYY565 pKa = 10.96DD566 pKa = 3.8YY567 pKa = 11.15MFNAANNNSDD577 pKa = 2.56HH578 pKa = 6.82FYY580 pKa = 10.99NVFSVGIKK588 pKa = 10.11ALRR591 pKa = 11.84PMEE594 pKa = 4.67TISKK598 pKa = 10.27ALEE601 pKa = 4.25TEE603 pKa = 4.13DD604 pKa = 3.74TNDD607 pKa = 2.91MGRR610 pKa = 11.84KK611 pKa = 9.22VRR613 pKa = 11.84VSMNN617 pKa = 3.02
Molecular weight: 70.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7I5|A0A4P8W7I5_9VIRU Helix-turn-helix XRE family-like protein OS=Capybara microvirus Cap1_SP_60 OX=2584785 PE=4 SV=1
MM1 pKa = 7.2CTNPKK6 pKa = 8.91TIKK9 pKa = 10.22NPKK12 pKa = 8.36IDD14 pKa = 3.74YY15 pKa = 9.97IKK17 pKa = 11.01YY18 pKa = 8.51YY19 pKa = 10.1DD20 pKa = 3.57KK21 pKa = 10.93PYY23 pKa = 10.49IQVPCGKK30 pKa = 10.08CKK32 pKa = 10.36EE33 pKa = 4.2CQEE36 pKa = 4.34LKK38 pKa = 10.28NNEE41 pKa = 3.72WLVRR45 pKa = 11.84NFFEE49 pKa = 4.37CKK51 pKa = 9.19EE52 pKa = 4.22AIRR55 pKa = 11.84CGGYY59 pKa = 7.09TQAYY63 pKa = 4.74TTTYY67 pKa = 9.45RR68 pKa = 11.84DD69 pKa = 2.69EE70 pKa = 6.1DD71 pKa = 3.57IPHH74 pKa = 6.51NSFGVKK80 pKa = 9.92CFSKK84 pKa = 10.83EE85 pKa = 3.88DD86 pKa = 3.23VRR88 pKa = 11.84KK89 pKa = 10.46YY90 pKa = 10.49IDD92 pKa = 4.54AIKK95 pKa = 10.61SDD97 pKa = 3.39LRR99 pKa = 11.84RR100 pKa = 11.84TFGAEE105 pKa = 3.84KK106 pKa = 10.38INGKK110 pKa = 9.54FKK112 pKa = 10.7YY113 pKa = 10.37YY114 pKa = 10.14MSFEE118 pKa = 4.31YY119 pKa = 10.61GSKK122 pKa = 10.25DD123 pKa = 2.63WYY125 pKa = 10.79FNKK128 pKa = 8.77KK129 pKa = 6.43TKK131 pKa = 10.09KK132 pKa = 8.95YY133 pKa = 8.91QQATEE138 pKa = 4.23RR139 pKa = 11.84PHH141 pKa = 6.7CHH143 pKa = 6.91FIFHH147 pKa = 6.38IQWKK151 pKa = 7.66CTLGEE156 pKa = 3.77RR157 pKa = 11.84CVIKK161 pKa = 10.71RR162 pKa = 11.84ILKK165 pKa = 8.29KK166 pKa = 9.85HH167 pKa = 4.74WVKK170 pKa = 11.29GDD172 pKa = 3.08ITYY175 pKa = 7.62TTNTNDD181 pKa = 4.07GIIDD185 pKa = 3.49EE186 pKa = 4.33RR187 pKa = 11.84AIAGGIGAVRR197 pKa = 11.84YY198 pKa = 7.18LTNYY202 pKa = 10.13LNEE205 pKa = 4.43EE206 pKa = 4.14KK207 pKa = 10.97NPIPKK212 pKa = 9.45EE213 pKa = 4.07VKK215 pKa = 9.71EE216 pKa = 4.16GCQRR220 pKa = 11.84IEE222 pKa = 3.93KK223 pKa = 9.95KK224 pKa = 10.31IKK226 pKa = 9.2FEE228 pKa = 3.62QRR230 pKa = 11.84YY231 pKa = 8.45IYY233 pKa = 10.89LMTAEE238 pKa = 4.95KK239 pKa = 9.31PQCEE243 pKa = 4.45SKK245 pKa = 10.46QEE247 pKa = 3.76KK248 pKa = 10.34AILYY252 pKa = 9.14KK253 pKa = 10.31VRR255 pKa = 11.84EE256 pKa = 4.16YY257 pKa = 11.68NNVVKK262 pKa = 10.37EE263 pKa = 4.12KK264 pKa = 10.39KK265 pKa = 9.74IKK267 pKa = 9.05NTEE270 pKa = 3.69EE271 pKa = 3.89RR272 pKa = 11.84IKK274 pKa = 10.43ILKK277 pKa = 9.63RR278 pKa = 11.84EE279 pKa = 3.87KK280 pKa = 10.53AKK282 pKa = 10.65CIPCTMQSSGLGKK295 pKa = 10.13SALLQDD301 pKa = 3.69QNGKK305 pKa = 10.24YY306 pKa = 9.94IYY308 pKa = 9.09NTYY311 pKa = 10.34FNYY314 pKa = 10.9AMAEE318 pKa = 3.96KK319 pKa = 10.34GEE321 pKa = 4.19CFIDD325 pKa = 4.68DD326 pKa = 4.31FEE328 pKa = 4.51LTRR331 pKa = 11.84KK332 pKa = 9.15KK333 pKa = 10.51RR334 pKa = 11.84KK335 pKa = 9.13LPLYY339 pKa = 10.02IEE341 pKa = 4.59RR342 pKa = 11.84KK343 pKa = 9.35LFYY346 pKa = 10.79KK347 pKa = 10.01EE348 pKa = 4.38VPCKK352 pKa = 9.89KK353 pKa = 9.83AKK355 pKa = 10.66NGMTYY360 pKa = 11.1KK361 pKa = 10.64LNEE364 pKa = 4.19AGKK367 pKa = 9.09EE368 pKa = 3.95MKK370 pKa = 10.07RR371 pKa = 11.84KK372 pKa = 9.54RR373 pKa = 11.84FHH375 pKa = 6.77FFALALQKK383 pKa = 10.69KK384 pKa = 7.97IWNVGHH390 pKa = 6.48YY391 pKa = 9.71DD392 pKa = 3.82YY393 pKa = 11.08NTMYY397 pKa = 10.28QYY399 pKa = 9.29KK400 pKa = 9.28TNEE403 pKa = 3.59EE404 pKa = 3.7KK405 pKa = 10.88AQYY408 pKa = 11.19KK409 pKa = 10.45KK410 pKa = 9.54MAKK413 pKa = 10.17DD414 pKa = 2.98ILANLKK420 pKa = 10.4DD421 pKa = 3.59FGMIEE426 pKa = 4.07RR427 pKa = 11.84YY428 pKa = 7.41VTYY431 pKa = 10.51KK432 pKa = 10.76GVEE435 pKa = 4.12KK436 pKa = 10.64KK437 pKa = 9.94DD438 pKa = 3.12TDD440 pKa = 3.66IINNYY445 pKa = 7.3GTLEE449 pKa = 4.19RR450 pKa = 11.84EE451 pKa = 4.8CQDD454 pKa = 2.52TWNILYY460 pKa = 9.95KK461 pKa = 9.89YY462 pKa = 9.59EE463 pKa = 4.13KK464 pKa = 10.77GKK466 pKa = 10.77EE467 pKa = 3.99EE468 pKa = 3.98EE469 pKa = 4.28NAEE472 pKa = 3.74QGYY475 pKa = 9.35KK476 pKa = 10.35FKK478 pKa = 11.23EE479 pKa = 3.99FLEE482 pKa = 5.02DD483 pKa = 3.98FAQGATSWGLMEE495 pKa = 4.22EE496 pKa = 5.11PIWQDD501 pKa = 3.16FVKK504 pKa = 10.72LSEE507 pKa = 4.23IYY509 pKa = 9.71TLEE512 pKa = 3.75KK513 pKa = 10.46TEE515 pKa = 4.09EE516 pKa = 3.99KK517 pKa = 10.7KK518 pKa = 10.99KK519 pKa = 9.57FDD521 pKa = 3.04RR522 pKa = 11.84MKK524 pKa = 10.64RR525 pKa = 11.84IRR527 pKa = 11.84QLHH530 pKa = 5.73KK531 pKa = 10.92KK532 pKa = 8.86RR533 pKa = 11.84MKK535 pKa = 10.59YY536 pKa = 9.61YY537 pKa = 10.72AA538 pKa = 4.15
MM1 pKa = 7.2CTNPKK6 pKa = 8.91TIKK9 pKa = 10.22NPKK12 pKa = 8.36IDD14 pKa = 3.74YY15 pKa = 9.97IKK17 pKa = 11.01YY18 pKa = 8.51YY19 pKa = 10.1DD20 pKa = 3.57KK21 pKa = 10.93PYY23 pKa = 10.49IQVPCGKK30 pKa = 10.08CKK32 pKa = 10.36EE33 pKa = 4.2CQEE36 pKa = 4.34LKK38 pKa = 10.28NNEE41 pKa = 3.72WLVRR45 pKa = 11.84NFFEE49 pKa = 4.37CKK51 pKa = 9.19EE52 pKa = 4.22AIRR55 pKa = 11.84CGGYY59 pKa = 7.09TQAYY63 pKa = 4.74TTTYY67 pKa = 9.45RR68 pKa = 11.84DD69 pKa = 2.69EE70 pKa = 6.1DD71 pKa = 3.57IPHH74 pKa = 6.51NSFGVKK80 pKa = 9.92CFSKK84 pKa = 10.83EE85 pKa = 3.88DD86 pKa = 3.23VRR88 pKa = 11.84KK89 pKa = 10.46YY90 pKa = 10.49IDD92 pKa = 4.54AIKK95 pKa = 10.61SDD97 pKa = 3.39LRR99 pKa = 11.84RR100 pKa = 11.84TFGAEE105 pKa = 3.84KK106 pKa = 10.38INGKK110 pKa = 9.54FKK112 pKa = 10.7YY113 pKa = 10.37YY114 pKa = 10.14MSFEE118 pKa = 4.31YY119 pKa = 10.61GSKK122 pKa = 10.25DD123 pKa = 2.63WYY125 pKa = 10.79FNKK128 pKa = 8.77KK129 pKa = 6.43TKK131 pKa = 10.09KK132 pKa = 8.95YY133 pKa = 8.91QQATEE138 pKa = 4.23RR139 pKa = 11.84PHH141 pKa = 6.7CHH143 pKa = 6.91FIFHH147 pKa = 6.38IQWKK151 pKa = 7.66CTLGEE156 pKa = 3.77RR157 pKa = 11.84CVIKK161 pKa = 10.71RR162 pKa = 11.84ILKK165 pKa = 8.29KK166 pKa = 9.85HH167 pKa = 4.74WVKK170 pKa = 11.29GDD172 pKa = 3.08ITYY175 pKa = 7.62TTNTNDD181 pKa = 4.07GIIDD185 pKa = 3.49EE186 pKa = 4.33RR187 pKa = 11.84AIAGGIGAVRR197 pKa = 11.84YY198 pKa = 7.18LTNYY202 pKa = 10.13LNEE205 pKa = 4.43EE206 pKa = 4.14KK207 pKa = 10.97NPIPKK212 pKa = 9.45EE213 pKa = 4.07VKK215 pKa = 9.71EE216 pKa = 4.16GCQRR220 pKa = 11.84IEE222 pKa = 3.93KK223 pKa = 9.95KK224 pKa = 10.31IKK226 pKa = 9.2FEE228 pKa = 3.62QRR230 pKa = 11.84YY231 pKa = 8.45IYY233 pKa = 10.89LMTAEE238 pKa = 4.95KK239 pKa = 9.31PQCEE243 pKa = 4.45SKK245 pKa = 10.46QEE247 pKa = 3.76KK248 pKa = 10.34AILYY252 pKa = 9.14KK253 pKa = 10.31VRR255 pKa = 11.84EE256 pKa = 4.16YY257 pKa = 11.68NNVVKK262 pKa = 10.37EE263 pKa = 4.12KK264 pKa = 10.39KK265 pKa = 9.74IKK267 pKa = 9.05NTEE270 pKa = 3.69EE271 pKa = 3.89RR272 pKa = 11.84IKK274 pKa = 10.43ILKK277 pKa = 9.63RR278 pKa = 11.84EE279 pKa = 3.87KK280 pKa = 10.53AKK282 pKa = 10.65CIPCTMQSSGLGKK295 pKa = 10.13SALLQDD301 pKa = 3.69QNGKK305 pKa = 10.24YY306 pKa = 9.94IYY308 pKa = 9.09NTYY311 pKa = 10.34FNYY314 pKa = 10.9AMAEE318 pKa = 3.96KK319 pKa = 10.34GEE321 pKa = 4.19CFIDD325 pKa = 4.68DD326 pKa = 4.31FEE328 pKa = 4.51LTRR331 pKa = 11.84KK332 pKa = 9.15KK333 pKa = 10.51RR334 pKa = 11.84KK335 pKa = 9.13LPLYY339 pKa = 10.02IEE341 pKa = 4.59RR342 pKa = 11.84KK343 pKa = 9.35LFYY346 pKa = 10.79KK347 pKa = 10.01EE348 pKa = 4.38VPCKK352 pKa = 9.89KK353 pKa = 9.83AKK355 pKa = 10.66NGMTYY360 pKa = 11.1KK361 pKa = 10.64LNEE364 pKa = 4.19AGKK367 pKa = 9.09EE368 pKa = 3.95MKK370 pKa = 10.07RR371 pKa = 11.84KK372 pKa = 9.54RR373 pKa = 11.84FHH375 pKa = 6.77FFALALQKK383 pKa = 10.69KK384 pKa = 7.97IWNVGHH390 pKa = 6.48YY391 pKa = 9.71DD392 pKa = 3.82YY393 pKa = 11.08NTMYY397 pKa = 10.28QYY399 pKa = 9.29KK400 pKa = 9.28TNEE403 pKa = 3.59EE404 pKa = 3.7KK405 pKa = 10.88AQYY408 pKa = 11.19KK409 pKa = 10.45KK410 pKa = 9.54MAKK413 pKa = 10.17DD414 pKa = 2.98ILANLKK420 pKa = 10.4DD421 pKa = 3.59FGMIEE426 pKa = 4.07RR427 pKa = 11.84YY428 pKa = 7.41VTYY431 pKa = 10.51KK432 pKa = 10.76GVEE435 pKa = 4.12KK436 pKa = 10.64KK437 pKa = 9.94DD438 pKa = 3.12TDD440 pKa = 3.66IINNYY445 pKa = 7.3GTLEE449 pKa = 4.19RR450 pKa = 11.84EE451 pKa = 4.8CQDD454 pKa = 2.52TWNILYY460 pKa = 9.95KK461 pKa = 9.89YY462 pKa = 9.59EE463 pKa = 4.13KK464 pKa = 10.77GKK466 pKa = 10.77EE467 pKa = 3.99EE468 pKa = 3.98EE469 pKa = 4.28NAEE472 pKa = 3.74QGYY475 pKa = 9.35KK476 pKa = 10.35FKK478 pKa = 11.23EE479 pKa = 3.99FLEE482 pKa = 5.02DD483 pKa = 3.98FAQGATSWGLMEE495 pKa = 4.22EE496 pKa = 5.11PIWQDD501 pKa = 3.16FVKK504 pKa = 10.72LSEE507 pKa = 4.23IYY509 pKa = 9.71TLEE512 pKa = 3.75KK513 pKa = 10.46TEE515 pKa = 4.09EE516 pKa = 3.99KK517 pKa = 10.7KK518 pKa = 10.99KK519 pKa = 9.57FDD521 pKa = 3.04RR522 pKa = 11.84MKK524 pKa = 10.64RR525 pKa = 11.84IRR527 pKa = 11.84QLHH530 pKa = 5.73KK531 pKa = 10.92KK532 pKa = 8.86RR533 pKa = 11.84MKK535 pKa = 10.59YY536 pKa = 9.61YY537 pKa = 10.72AA538 pKa = 4.15
Molecular weight: 64.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1725 |
94 |
617 |
345.0 |
39.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.246 ± 1.83 | 1.681 ± 0.59 |
4.696 ± 0.585 | 8.348 ± 1.092 |
4.406 ± 0.799 | 6.261 ± 0.78 |
1.159 ± 0.444 | 7.826 ± 0.36 |
9.159 ± 2.598 | 6.087 ± 0.469 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.536 ± 0.409 | 7.536 ± 0.916 |
2.725 ± 0.51 | 5.043 ± 1.037 |
4.232 ± 0.461 | 4.464 ± 1.058 |
6.029 ± 0.411 | 3.246 ± 0.326 |
1.681 ± 0.309 | 4.638 ± 1.23 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
