
Myroides profundi
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Myroides
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
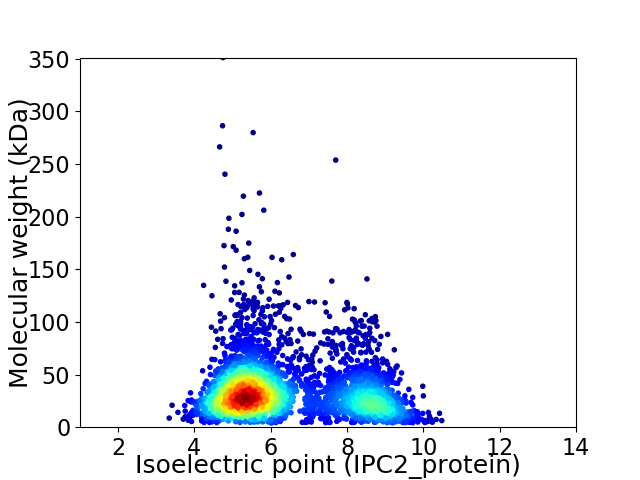
Virtual 2D-PAGE plot for 3534 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5RHP8|A0A0B5RHP8_9FLAO Peptidase S9 OS=Myroides profundi OX=480520 GN=MPR_1081 PE=4 SV=1
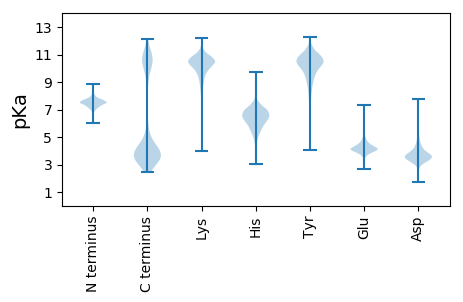
MM1 pKa = 7.69KK2 pKa = 10.74LNTTNRR8 pKa = 11.84ADD10 pKa = 3.82LPNPKK15 pKa = 9.12ILKK18 pKa = 8.47EE19 pKa = 4.04LCKK22 pKa = 10.74SLATLDD28 pKa = 5.69AILCQDD34 pKa = 3.0WEE36 pKa = 4.24YY37 pKa = 11.07RR38 pKa = 11.84YY39 pKa = 10.92YY40 pKa = 10.99LYY42 pKa = 11.38NHH44 pKa = 7.2LWSDD48 pKa = 3.84DD49 pKa = 3.58EE50 pKa = 5.68EE51 pKa = 4.48FFQMRR56 pKa = 11.84DD57 pKa = 3.23GEE59 pKa = 4.5GNDD62 pKa = 3.25MVILFRR68 pKa = 11.84VDD70 pKa = 2.96GTVINGFDD78 pKa = 3.25HH79 pKa = 6.74EE80 pKa = 5.18LYY82 pKa = 10.74DD83 pKa = 5.13FEE85 pKa = 5.85EE86 pKa = 4.2NLPNKK91 pKa = 10.0EE92 pKa = 4.46DD93 pKa = 3.66LTAGLPNQYY102 pKa = 9.92IEE104 pKa = 5.23FIFGEE109 pKa = 4.64YY110 pKa = 9.83ISDD113 pKa = 3.62IGTTYY118 pKa = 10.64CIWSDD123 pKa = 4.63NNQQWAIGQVDD134 pKa = 3.69NEE136 pKa = 4.46EE137 pKa = 5.0DD138 pKa = 3.82GSDD141 pKa = 3.45DD142 pKa = 3.91QLYY145 pKa = 10.47IFDD148 pKa = 5.15NNPDD152 pKa = 3.65TYY154 pKa = 11.22INWATNYY161 pKa = 10.15YY162 pKa = 9.99FDD164 pKa = 4.68EE165 pKa = 4.83EE166 pKa = 4.67EE167 pKa = 4.26EE168 pKa = 4.95SMTADD173 pKa = 3.0KK174 pKa = 11.0KK175 pKa = 11.16DD176 pKa = 3.83IIRR179 pKa = 11.84QIYY182 pKa = 8.22QGKK185 pKa = 7.27TLTKK189 pKa = 10.79EE190 pKa = 4.19MILTLNPEE198 pKa = 4.18LEE200 pKa = 4.1DD201 pKa = 3.64WEE203 pKa = 4.53LLKK206 pKa = 10.94EE207 pKa = 4.59DD208 pKa = 4.59IIEE211 pKa = 3.98INYY214 pKa = 8.21PHH216 pKa = 6.86SLL218 pKa = 3.5
MM1 pKa = 7.69KK2 pKa = 10.74LNTTNRR8 pKa = 11.84ADD10 pKa = 3.82LPNPKK15 pKa = 9.12ILKK18 pKa = 8.47EE19 pKa = 4.04LCKK22 pKa = 10.74SLATLDD28 pKa = 5.69AILCQDD34 pKa = 3.0WEE36 pKa = 4.24YY37 pKa = 11.07RR38 pKa = 11.84YY39 pKa = 10.92YY40 pKa = 10.99LYY42 pKa = 11.38NHH44 pKa = 7.2LWSDD48 pKa = 3.84DD49 pKa = 3.58EE50 pKa = 5.68EE51 pKa = 4.48FFQMRR56 pKa = 11.84DD57 pKa = 3.23GEE59 pKa = 4.5GNDD62 pKa = 3.25MVILFRR68 pKa = 11.84VDD70 pKa = 2.96GTVINGFDD78 pKa = 3.25HH79 pKa = 6.74EE80 pKa = 5.18LYY82 pKa = 10.74DD83 pKa = 5.13FEE85 pKa = 5.85EE86 pKa = 4.2NLPNKK91 pKa = 10.0EE92 pKa = 4.46DD93 pKa = 3.66LTAGLPNQYY102 pKa = 9.92IEE104 pKa = 5.23FIFGEE109 pKa = 4.64YY110 pKa = 9.83ISDD113 pKa = 3.62IGTTYY118 pKa = 10.64CIWSDD123 pKa = 4.63NNQQWAIGQVDD134 pKa = 3.69NEE136 pKa = 4.46EE137 pKa = 5.0DD138 pKa = 3.82GSDD141 pKa = 3.45DD142 pKa = 3.91QLYY145 pKa = 10.47IFDD148 pKa = 5.15NNPDD152 pKa = 3.65TYY154 pKa = 11.22INWATNYY161 pKa = 10.15YY162 pKa = 9.99FDD164 pKa = 4.68EE165 pKa = 4.83EE166 pKa = 4.67EE167 pKa = 4.26EE168 pKa = 4.95SMTADD173 pKa = 3.0KK174 pKa = 11.0KK175 pKa = 11.16DD176 pKa = 3.83IIRR179 pKa = 11.84QIYY182 pKa = 8.22QGKK185 pKa = 7.27TLTKK189 pKa = 10.79EE190 pKa = 4.19MILTLNPEE198 pKa = 4.18LEE200 pKa = 4.1DD201 pKa = 3.64WEE203 pKa = 4.53LLKK206 pKa = 10.94EE207 pKa = 4.59DD208 pKa = 4.59IIEE211 pKa = 3.98INYY214 pKa = 8.21PHH216 pKa = 6.86SLL218 pKa = 3.5
Molecular weight: 25.83 kDa
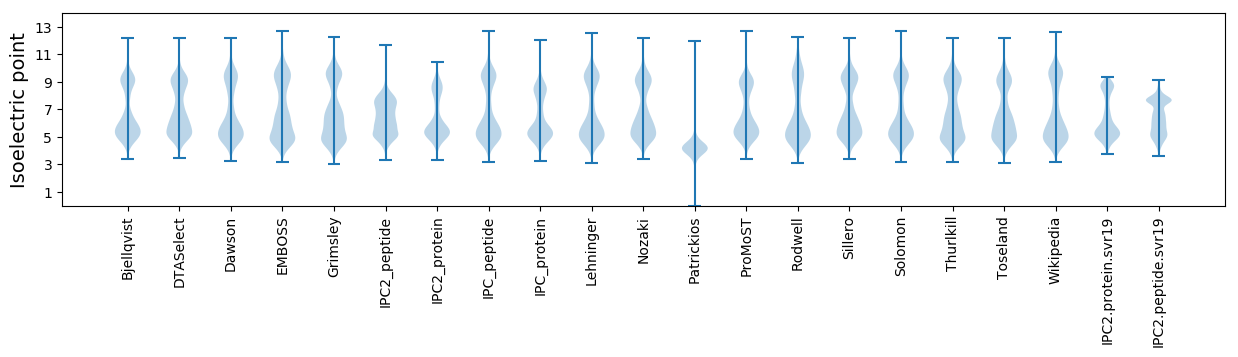
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5RWD5|A0A0B5RWD5_9FLAO Mechanosensitive ion channel protein MscS OS=Myroides profundi OX=480520 GN=MPR_1159 PE=4 SV=1
MM1 pKa = 8.03IEE3 pKa = 4.28DD4 pKa = 4.01VIYY7 pKa = 9.97NQWSIILGLLYY18 pKa = 10.3TGWLTSRR25 pKa = 11.84IVKK28 pKa = 8.88RR29 pKa = 11.84TTKK32 pKa = 9.96GRR34 pKa = 11.84KK35 pKa = 8.21EE36 pKa = 3.56RR37 pKa = 11.84AGCVDD42 pKa = 4.06VIRR45 pKa = 11.84VFLIIGGLFGLTVFGSVAFGSMFIRR70 pKa = 11.84TLSSLINGTVVFSIEE85 pKa = 3.87LFAEE89 pKa = 4.69LVCYY93 pKa = 9.18GTMVAVLVFAFIGVMLYY110 pKa = 10.84GVGRR114 pKa = 11.84DD115 pKa = 3.14MRR117 pKa = 11.84RR118 pKa = 11.84YY119 pKa = 9.54NRR121 pKa = 11.84LARR124 pKa = 11.84LLGLGVLVPLIMLVFEE140 pKa = 4.65GALVYY145 pKa = 10.96ALVRR149 pKa = 11.84NIHH152 pKa = 6.4NIPLSVLLTVFILILLIGIILYY174 pKa = 9.86VKK176 pKa = 10.17IIRR179 pKa = 11.84EE180 pKa = 4.27EE181 pKa = 3.77ITDD184 pKa = 4.79LIEE187 pKa = 4.18KK188 pKa = 9.12EE189 pKa = 4.37KK190 pKa = 10.8NASRR194 pKa = 11.84SKK196 pKa = 10.02EE197 pKa = 3.93NKK199 pKa = 8.75TKK201 pKa = 11.03GITQDD206 pKa = 3.11KK207 pKa = 10.36SVTRR211 pKa = 11.84AQTANHH217 pKa = 5.98SRR219 pKa = 11.84QEE221 pKa = 4.5SNRR224 pKa = 11.84STQRR228 pKa = 11.84TNKK231 pKa = 9.37PNPHH235 pKa = 5.36NDD237 pKa = 2.57EE238 pKa = 3.72WKK240 pKa = 9.99RR241 pKa = 11.84RR242 pKa = 11.84SNRR245 pKa = 11.84RR246 pKa = 11.84KK247 pKa = 9.98KK248 pKa = 10.38RR249 pKa = 3.33
MM1 pKa = 8.03IEE3 pKa = 4.28DD4 pKa = 4.01VIYY7 pKa = 9.97NQWSIILGLLYY18 pKa = 10.3TGWLTSRR25 pKa = 11.84IVKK28 pKa = 8.88RR29 pKa = 11.84TTKK32 pKa = 9.96GRR34 pKa = 11.84KK35 pKa = 8.21EE36 pKa = 3.56RR37 pKa = 11.84AGCVDD42 pKa = 4.06VIRR45 pKa = 11.84VFLIIGGLFGLTVFGSVAFGSMFIRR70 pKa = 11.84TLSSLINGTVVFSIEE85 pKa = 3.87LFAEE89 pKa = 4.69LVCYY93 pKa = 9.18GTMVAVLVFAFIGVMLYY110 pKa = 10.84GVGRR114 pKa = 11.84DD115 pKa = 3.14MRR117 pKa = 11.84RR118 pKa = 11.84YY119 pKa = 9.54NRR121 pKa = 11.84LARR124 pKa = 11.84LLGLGVLVPLIMLVFEE140 pKa = 4.65GALVYY145 pKa = 10.96ALVRR149 pKa = 11.84NIHH152 pKa = 6.4NIPLSVLLTVFILILLIGIILYY174 pKa = 9.86VKK176 pKa = 10.17IIRR179 pKa = 11.84EE180 pKa = 4.27EE181 pKa = 3.77ITDD184 pKa = 4.79LIEE187 pKa = 4.18KK188 pKa = 9.12EE189 pKa = 4.37KK190 pKa = 10.8NASRR194 pKa = 11.84SKK196 pKa = 10.02EE197 pKa = 3.93NKK199 pKa = 8.75TKK201 pKa = 11.03GITQDD206 pKa = 3.11KK207 pKa = 10.36SVTRR211 pKa = 11.84AQTANHH217 pKa = 5.98SRR219 pKa = 11.84QEE221 pKa = 4.5SNRR224 pKa = 11.84STQRR228 pKa = 11.84TNKK231 pKa = 9.37PNPHH235 pKa = 5.36NDD237 pKa = 2.57EE238 pKa = 3.72WKK240 pKa = 9.99RR241 pKa = 11.84RR242 pKa = 11.84SNRR245 pKa = 11.84RR246 pKa = 11.84KK247 pKa = 9.98KK248 pKa = 10.38RR249 pKa = 3.33
Molecular weight: 28.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1144917 |
37 |
3339 |
324.0 |
36.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.181 ± 0.049 | 0.727 ± 0.013 |
5.552 ± 0.032 | 6.822 ± 0.048 |
4.913 ± 0.033 | 6.246 ± 0.046 |
1.755 ± 0.021 | 7.968 ± 0.038 |
7.839 ± 0.04 | 9.449 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.338 ± 0.025 | 5.783 ± 0.043 |
3.125 ± 0.022 | 3.542 ± 0.025 |
3.39 ± 0.03 | 6.343 ± 0.035 |
5.798 ± 0.044 | 6.702 ± 0.038 |
0.975 ± 0.014 | 4.552 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
