
Macaca nemestrina herpesvirus 7
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Betaherpesvirinae; Roseolovirus; Macacine betaherpesvirus 9
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
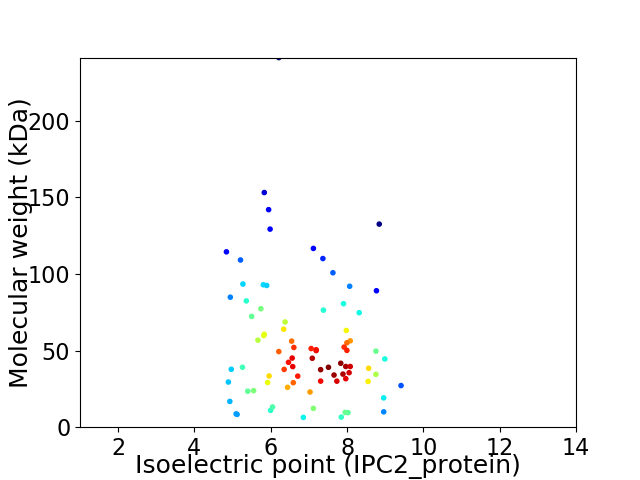
Virtual 2D-PAGE plot for 83 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
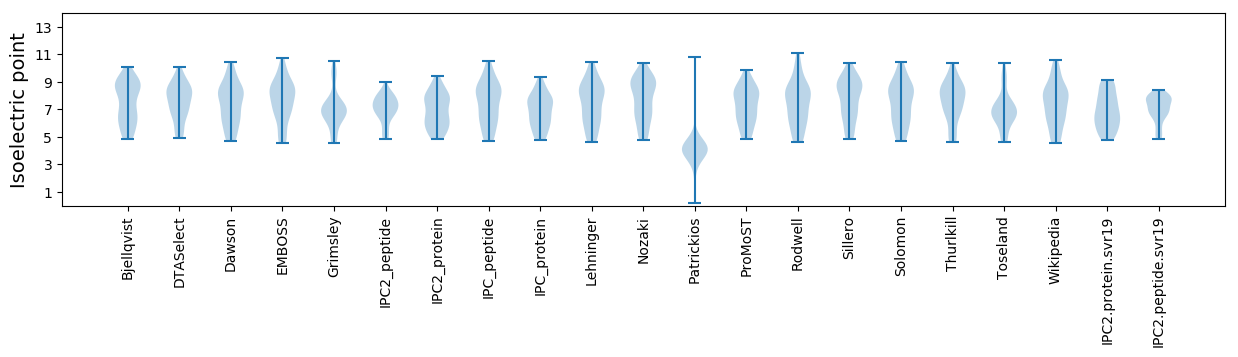
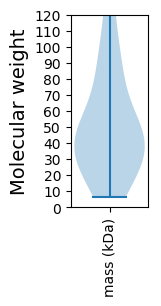
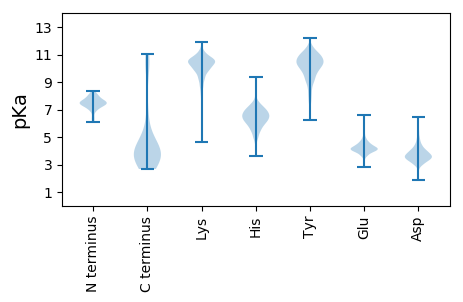
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A191S3V9|A0A191S3V9_9BETA Uracil-DNA glycosylase OS=Macaca nemestrina herpesvirus 7 OX=1846169 GN=U81 PE=3 SV=1
MM1 pKa = 6.23QTNIYY6 pKa = 10.37FCFFFNFSTSLYY18 pKa = 10.68ALFNNSYY25 pKa = 10.47YY26 pKa = 9.78STLGKK31 pKa = 10.45EE32 pKa = 4.79CINAIKK38 pKa = 10.63NCTQSEE44 pKa = 4.35RR45 pKa = 11.84PILLEE50 pKa = 5.52PIDD53 pKa = 4.15QAPTPNPDD61 pKa = 3.53VISGLLYY68 pKa = 7.12YY69 pKa = 9.99TPYY72 pKa = 11.03SFDD75 pKa = 3.52EE76 pKa = 4.34YY77 pKa = 11.08QVLVDD82 pKa = 4.79EE83 pKa = 5.05EE84 pKa = 4.42FLNTFYY90 pKa = 11.65LLYY93 pKa = 10.75NNPNQLHH100 pKa = 5.71VLFSLIKK107 pKa = 9.77DD108 pKa = 3.95SKK110 pKa = 11.25SKK112 pKa = 10.97SSWLGFLNSFEE123 pKa = 4.81KK124 pKa = 10.87CFSDD128 pKa = 3.81NTIITCRR135 pKa = 11.84NQVCKK140 pKa = 9.96TYY142 pKa = 10.52SYY144 pKa = 11.45EE145 pKa = 4.02NLKK148 pKa = 8.55YY149 pKa = 8.93TPNIFVEE156 pKa = 4.43NIIGFEE162 pKa = 4.06FYY164 pKa = 10.35ISEE167 pKa = 4.27NNLDD171 pKa = 3.44VSFNSSILIYY181 pKa = 10.17LQNEE185 pKa = 3.66EE186 pKa = 4.0TRR188 pKa = 11.84AKK190 pKa = 10.33KK191 pKa = 9.53IVKK194 pKa = 9.73IMYY197 pKa = 9.53EE198 pKa = 4.28GINVFDD204 pKa = 4.72ALLNSMRR211 pKa = 11.84YY212 pKa = 9.4FSQKK216 pKa = 9.96FQFSFDD222 pKa = 3.67FPLLRR227 pKa = 11.84EE228 pKa = 4.08MEE230 pKa = 4.58SYY232 pKa = 11.62NDD234 pKa = 3.8LLPFRR239 pKa = 11.84SEE241 pKa = 3.87PSNLLIRR248 pKa = 11.84THH250 pKa = 6.33
MM1 pKa = 6.23QTNIYY6 pKa = 10.37FCFFFNFSTSLYY18 pKa = 10.68ALFNNSYY25 pKa = 10.47YY26 pKa = 9.78STLGKK31 pKa = 10.45EE32 pKa = 4.79CINAIKK38 pKa = 10.63NCTQSEE44 pKa = 4.35RR45 pKa = 11.84PILLEE50 pKa = 5.52PIDD53 pKa = 4.15QAPTPNPDD61 pKa = 3.53VISGLLYY68 pKa = 7.12YY69 pKa = 9.99TPYY72 pKa = 11.03SFDD75 pKa = 3.52EE76 pKa = 4.34YY77 pKa = 11.08QVLVDD82 pKa = 4.79EE83 pKa = 5.05EE84 pKa = 4.42FLNTFYY90 pKa = 11.65LLYY93 pKa = 10.75NNPNQLHH100 pKa = 5.71VLFSLIKK107 pKa = 9.77DD108 pKa = 3.95SKK110 pKa = 11.25SKK112 pKa = 10.97SSWLGFLNSFEE123 pKa = 4.81KK124 pKa = 10.87CFSDD128 pKa = 3.81NTIITCRR135 pKa = 11.84NQVCKK140 pKa = 9.96TYY142 pKa = 10.52SYY144 pKa = 11.45EE145 pKa = 4.02NLKK148 pKa = 8.55YY149 pKa = 8.93TPNIFVEE156 pKa = 4.43NIIGFEE162 pKa = 4.06FYY164 pKa = 10.35ISEE167 pKa = 4.27NNLDD171 pKa = 3.44VSFNSSILIYY181 pKa = 10.17LQNEE185 pKa = 3.66EE186 pKa = 4.0TRR188 pKa = 11.84AKK190 pKa = 10.33KK191 pKa = 9.53IVKK194 pKa = 9.73IMYY197 pKa = 9.53EE198 pKa = 4.28GINVFDD204 pKa = 4.72ALLNSMRR211 pKa = 11.84YY212 pKa = 9.4FSQKK216 pKa = 9.96FQFSFDD222 pKa = 3.67FPLLRR227 pKa = 11.84EE228 pKa = 4.08MEE230 pKa = 4.58SYY232 pKa = 11.62NDD234 pKa = 3.8LLPFRR239 pKa = 11.84SEE241 pKa = 3.87PSNLLIRR248 pKa = 11.84THH250 pKa = 6.33
Molecular weight: 29.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A192XPA1|A0A192XPA1_9BETA Capsid scaffold protein OS=Macaca nemestrina herpesvirus 7 OX=1846169 GN=U53.5 PE=4 SV=1
MM1 pKa = 7.68SLEE4 pKa = 4.18HH5 pKa = 6.66LPAIRR10 pKa = 11.84KK11 pKa = 9.48CIGQYY16 pKa = 9.19NHH18 pKa = 6.03LRR20 pKa = 11.84IYY22 pKa = 10.87KK23 pKa = 9.63KK24 pKa = 10.42ILMLKK29 pKa = 10.66SNFTEE34 pKa = 4.42LNFFLGNIFPHH45 pKa = 6.58EE46 pKa = 4.05LQTSKK51 pKa = 10.16IHH53 pKa = 6.12VFFEE57 pKa = 4.22VTLGNRR63 pKa = 11.84IPDD66 pKa = 4.28CIIVLNHH73 pKa = 6.05LNEE76 pKa = 3.89NRR78 pKa = 11.84VNEE81 pKa = 3.53IHH83 pKa = 7.42LYY85 pKa = 9.69FFEE88 pKa = 5.1FKK90 pKa = 8.42TTFAKK95 pKa = 9.49STLFSIQKK103 pKa = 8.31NTTQKK108 pKa = 10.63LQYY111 pKa = 10.46LQGLKK116 pKa = 9.92QLQEE120 pKa = 3.66ATKK123 pKa = 10.42FLEE126 pKa = 3.92QYY128 pKa = 10.18LIKK131 pKa = 10.96NEE133 pKa = 4.3TTCKK137 pKa = 9.4ISPVICFFRR146 pKa = 11.84QRR148 pKa = 11.84GLKK151 pKa = 10.08LDD153 pKa = 3.8FVKK156 pKa = 10.3TFVSKK161 pKa = 10.42EE162 pKa = 4.06LQLSLPFILNLLAQFEE178 pKa = 4.54NDD180 pKa = 3.46TVKK183 pKa = 10.88SILSISDD190 pKa = 3.17TAHH193 pKa = 6.78FRR195 pKa = 11.84RR196 pKa = 11.84ACQKK200 pKa = 9.88YY201 pKa = 8.52SHH203 pKa = 6.29LQRR206 pKa = 11.84RR207 pKa = 11.84RR208 pKa = 11.84NSKK211 pKa = 9.21TSTNRR216 pKa = 11.84SNNTAKK222 pKa = 10.71KK223 pKa = 9.97KK224 pKa = 10.16RR225 pKa = 11.84RR226 pKa = 11.84NPKK229 pKa = 9.65KK230 pKa = 10.49
MM1 pKa = 7.68SLEE4 pKa = 4.18HH5 pKa = 6.66LPAIRR10 pKa = 11.84KK11 pKa = 9.48CIGQYY16 pKa = 9.19NHH18 pKa = 6.03LRR20 pKa = 11.84IYY22 pKa = 10.87KK23 pKa = 9.63KK24 pKa = 10.42ILMLKK29 pKa = 10.66SNFTEE34 pKa = 4.42LNFFLGNIFPHH45 pKa = 6.58EE46 pKa = 4.05LQTSKK51 pKa = 10.16IHH53 pKa = 6.12VFFEE57 pKa = 4.22VTLGNRR63 pKa = 11.84IPDD66 pKa = 4.28CIIVLNHH73 pKa = 6.05LNEE76 pKa = 3.89NRR78 pKa = 11.84VNEE81 pKa = 3.53IHH83 pKa = 7.42LYY85 pKa = 9.69FFEE88 pKa = 5.1FKK90 pKa = 8.42TTFAKK95 pKa = 9.49STLFSIQKK103 pKa = 8.31NTTQKK108 pKa = 10.63LQYY111 pKa = 10.46LQGLKK116 pKa = 9.92QLQEE120 pKa = 3.66ATKK123 pKa = 10.42FLEE126 pKa = 3.92QYY128 pKa = 10.18LIKK131 pKa = 10.96NEE133 pKa = 4.3TTCKK137 pKa = 9.4ISPVICFFRR146 pKa = 11.84QRR148 pKa = 11.84GLKK151 pKa = 10.08LDD153 pKa = 3.8FVKK156 pKa = 10.3TFVSKK161 pKa = 10.42EE162 pKa = 4.06LQLSLPFILNLLAQFEE178 pKa = 4.54NDD180 pKa = 3.46TVKK183 pKa = 10.88SILSISDD190 pKa = 3.17TAHH193 pKa = 6.78FRR195 pKa = 11.84RR196 pKa = 11.84ACQKK200 pKa = 9.88YY201 pKa = 8.52SHH203 pKa = 6.29LQRR206 pKa = 11.84RR207 pKa = 11.84RR208 pKa = 11.84NSKK211 pKa = 9.21TSTNRR216 pKa = 11.84SNNTAKK222 pKa = 10.71KK223 pKa = 9.97KK224 pKa = 10.16RR225 pKa = 11.84RR226 pKa = 11.84NPKK229 pKa = 9.65KK230 pKa = 10.49
Molecular weight: 27.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
39259 |
54 |
2056 |
473.0 |
54.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.584 ± 0.135 | 2.455 ± 0.164 |
4.791 ± 0.145 | 6.106 ± 0.197 |
6.371 ± 0.198 | 3.169 ± 0.172 |
2.394 ± 0.08 | 8.559 ± 0.244 |
7.496 ± 0.236 | 10.479 ± 0.226 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.198 ± 0.084 | 7.181 ± 0.197 |
3.505 ± 0.152 | 3.747 ± 0.15 |
3.691 ± 0.209 | 7.996 ± 0.248 |
6.218 ± 0.183 | 5.24 ± 0.155 |
0.769 ± 0.059 | 4.053 ± 0.123 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
