
Streptococcus satellite phage Javan114
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
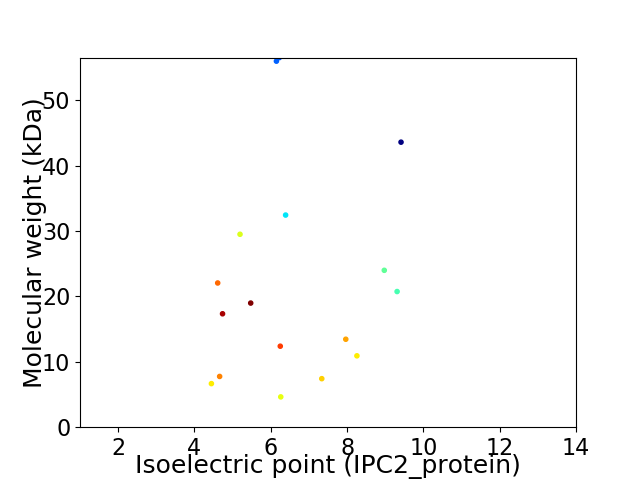
Virtual 2D-PAGE plot for 17 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
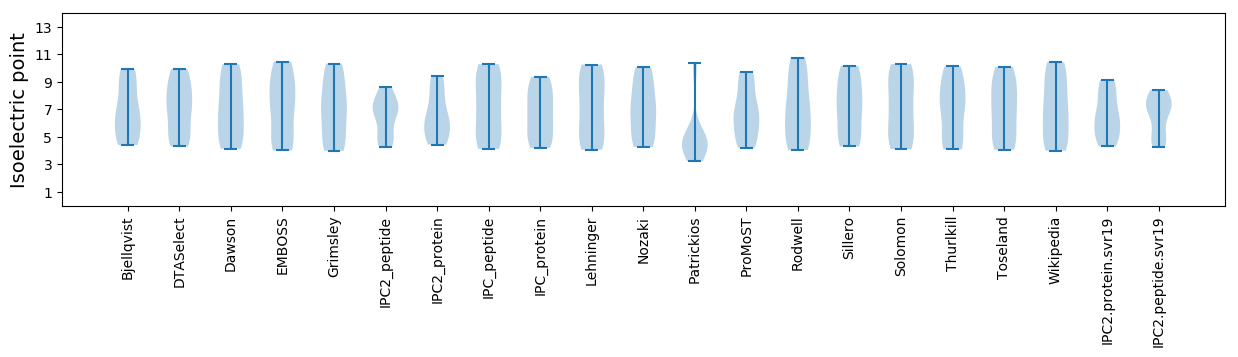
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZDH5|A0A4D5ZDH5_9VIRU Putative transcriptional regulator OS=Streptococcus satellite phage Javan114 OX=2558528 GN=JavanS114_0016 PE=4 SV=1
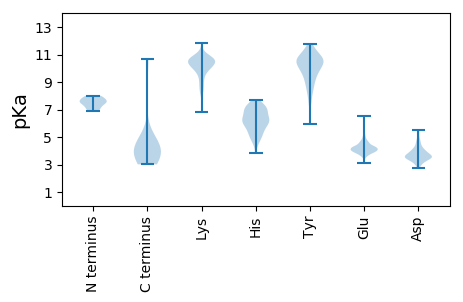
MM1 pKa = 7.69KK2 pKa = 10.27IKK4 pKa = 10.3IFYY7 pKa = 8.32QQQNQSLQEE16 pKa = 4.0FEE18 pKa = 4.69AQINNFMASVEE29 pKa = 4.24VVDD32 pKa = 5.09VKK34 pKa = 10.66CTEE37 pKa = 4.01ATEE40 pKa = 4.13GDD42 pKa = 4.36YY43 pKa = 11.64EE44 pKa = 4.22NLSATLGLLVLYY56 pKa = 10.69KK57 pKa = 10.48EE58 pKa = 4.5
MM1 pKa = 7.69KK2 pKa = 10.27IKK4 pKa = 10.3IFYY7 pKa = 8.32QQQNQSLQEE16 pKa = 4.0FEE18 pKa = 4.69AQINNFMASVEE29 pKa = 4.24VVDD32 pKa = 5.09VKK34 pKa = 10.66CTEE37 pKa = 4.01ATEE40 pKa = 4.13GDD42 pKa = 4.36YY43 pKa = 11.64EE44 pKa = 4.22NLSATLGLLVLYY56 pKa = 10.69KK57 pKa = 10.48EE58 pKa = 4.5
Molecular weight: 6.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZFT1|A0A4D5ZFT1_9VIRU PriCT_1 domain-containing protein OS=Streptococcus satellite phage Javan114 OX=2558528 GN=JavanS114_0008 PE=4 SV=1
MM1 pKa = 7.88KK2 pKa = 9.33ITEE5 pKa = 4.22VKK7 pKa = 10.53KK8 pKa = 10.48KK9 pKa = 10.5DD10 pKa = 3.31GSNVYY15 pKa = 9.72RR16 pKa = 11.84ASVYY20 pKa = 10.78LGVDD24 pKa = 2.9QVTGKK29 pKa = 10.15KK30 pKa = 10.12VKK32 pKa = 9.8TSITGRR38 pKa = 11.84TRR40 pKa = 11.84KK41 pKa = 8.72EE42 pKa = 4.07VKK44 pKa = 10.28SKK46 pKa = 10.67AQHH49 pKa = 5.72AQLDD53 pKa = 4.67FKK55 pKa = 11.77ANGSTVHH62 pKa = 5.11KK63 pKa = 8.56TVRR66 pKa = 11.84VKK68 pKa = 10.74NYY70 pKa = 10.28QEE72 pKa = 4.72LAEE75 pKa = 4.75LWLEE79 pKa = 4.21SYY81 pKa = 10.9RR82 pKa = 11.84LTVKK86 pKa = 10.03PQTFIATEE94 pKa = 3.93RR95 pKa = 11.84MVNSHH100 pKa = 7.68LIPVFGKK107 pKa = 9.06MRR109 pKa = 11.84LDD111 pKa = 3.4KK112 pKa = 10.95LAASYY117 pKa = 10.16IQSFINDD124 pKa = 3.53LSSRR128 pKa = 11.84LVHH131 pKa = 6.43FGVVHH136 pKa = 6.77SINSRR141 pKa = 11.84ILKK144 pKa = 10.27YY145 pKa = 10.48GVSLQLLPFNPARR158 pKa = 11.84DD159 pKa = 3.52IILPRR164 pKa = 11.84KK165 pKa = 8.83PKK167 pKa = 10.33RR168 pKa = 11.84EE169 pKa = 3.83SKK171 pKa = 10.3AIKK174 pKa = 10.18FIAPEE179 pKa = 4.14DD180 pKa = 3.8LKK182 pKa = 11.58SLLGYY187 pKa = 8.07MEE189 pKa = 4.55KK190 pKa = 10.67LAFKK194 pKa = 10.28KK195 pKa = 10.07YY196 pKa = 9.29SHH198 pKa = 6.33YY199 pKa = 10.57FDD201 pKa = 4.03YY202 pKa = 11.26VLYY205 pKa = 10.72SLLLATGCRR214 pKa = 11.84FGEE217 pKa = 4.29AVALEE222 pKa = 4.2WSDD225 pKa = 3.65IDD227 pKa = 5.39LEE229 pKa = 4.22NATISISKK237 pKa = 10.02NYY239 pKa = 9.17NRR241 pKa = 11.84LVDD244 pKa = 4.31LVGPPKK250 pKa = 10.52SKK252 pKa = 10.78AGVRR256 pKa = 11.84VISIDD261 pKa = 3.3RR262 pKa = 11.84KK263 pKa = 7.22TVNMLKK269 pKa = 10.23LYY271 pKa = 10.65KK272 pKa = 10.14NRR274 pKa = 11.84QRR276 pKa = 11.84QLFVEE281 pKa = 4.94DD282 pKa = 4.01GARR285 pKa = 11.84VPSVVFAIPTKK296 pKa = 10.24AYY298 pKa = 10.59QNMATRR304 pKa = 11.84QDD306 pKa = 3.65SLDD309 pKa = 3.68RR310 pKa = 11.84RR311 pKa = 11.84CKK313 pKa = 9.94EE314 pKa = 3.14IRR316 pKa = 11.84IPRR319 pKa = 11.84FTFHH323 pKa = 7.65AFRR326 pKa = 11.84HH327 pKa = 4.69THH329 pKa = 6.92ASLLLNAGISYY340 pKa = 10.68KK341 pKa = 10.23EE342 pKa = 3.61LQYY345 pKa = 11.49RR346 pKa = 11.84LGHH349 pKa = 6.19ATLAMTMDD357 pKa = 4.24IYY359 pKa = 11.79SHH361 pKa = 6.95LSKK364 pKa = 10.92DD365 pKa = 3.51KK366 pKa = 10.41EE367 pKa = 4.25KK368 pKa = 10.93EE369 pKa = 3.85AVSYY373 pKa = 9.52YY374 pKa = 10.59EE375 pKa = 4.03KK376 pKa = 10.7AINGLL381 pKa = 3.67
MM1 pKa = 7.88KK2 pKa = 9.33ITEE5 pKa = 4.22VKK7 pKa = 10.53KK8 pKa = 10.48KK9 pKa = 10.5DD10 pKa = 3.31GSNVYY15 pKa = 9.72RR16 pKa = 11.84ASVYY20 pKa = 10.78LGVDD24 pKa = 2.9QVTGKK29 pKa = 10.15KK30 pKa = 10.12VKK32 pKa = 9.8TSITGRR38 pKa = 11.84TRR40 pKa = 11.84KK41 pKa = 8.72EE42 pKa = 4.07VKK44 pKa = 10.28SKK46 pKa = 10.67AQHH49 pKa = 5.72AQLDD53 pKa = 4.67FKK55 pKa = 11.77ANGSTVHH62 pKa = 5.11KK63 pKa = 8.56TVRR66 pKa = 11.84VKK68 pKa = 10.74NYY70 pKa = 10.28QEE72 pKa = 4.72LAEE75 pKa = 4.75LWLEE79 pKa = 4.21SYY81 pKa = 10.9RR82 pKa = 11.84LTVKK86 pKa = 10.03PQTFIATEE94 pKa = 3.93RR95 pKa = 11.84MVNSHH100 pKa = 7.68LIPVFGKK107 pKa = 9.06MRR109 pKa = 11.84LDD111 pKa = 3.4KK112 pKa = 10.95LAASYY117 pKa = 10.16IQSFINDD124 pKa = 3.53LSSRR128 pKa = 11.84LVHH131 pKa = 6.43FGVVHH136 pKa = 6.77SINSRR141 pKa = 11.84ILKK144 pKa = 10.27YY145 pKa = 10.48GVSLQLLPFNPARR158 pKa = 11.84DD159 pKa = 3.52IILPRR164 pKa = 11.84KK165 pKa = 8.83PKK167 pKa = 10.33RR168 pKa = 11.84EE169 pKa = 3.83SKK171 pKa = 10.3AIKK174 pKa = 10.18FIAPEE179 pKa = 4.14DD180 pKa = 3.8LKK182 pKa = 11.58SLLGYY187 pKa = 8.07MEE189 pKa = 4.55KK190 pKa = 10.67LAFKK194 pKa = 10.28KK195 pKa = 10.07YY196 pKa = 9.29SHH198 pKa = 6.33YY199 pKa = 10.57FDD201 pKa = 4.03YY202 pKa = 11.26VLYY205 pKa = 10.72SLLLATGCRR214 pKa = 11.84FGEE217 pKa = 4.29AVALEE222 pKa = 4.2WSDD225 pKa = 3.65IDD227 pKa = 5.39LEE229 pKa = 4.22NATISISKK237 pKa = 10.02NYY239 pKa = 9.17NRR241 pKa = 11.84LVDD244 pKa = 4.31LVGPPKK250 pKa = 10.52SKK252 pKa = 10.78AGVRR256 pKa = 11.84VISIDD261 pKa = 3.3RR262 pKa = 11.84KK263 pKa = 7.22TVNMLKK269 pKa = 10.23LYY271 pKa = 10.65KK272 pKa = 10.14NRR274 pKa = 11.84QRR276 pKa = 11.84QLFVEE281 pKa = 4.94DD282 pKa = 4.01GARR285 pKa = 11.84VPSVVFAIPTKK296 pKa = 10.24AYY298 pKa = 10.59QNMATRR304 pKa = 11.84QDD306 pKa = 3.65SLDD309 pKa = 3.68RR310 pKa = 11.84RR311 pKa = 11.84CKK313 pKa = 9.94EE314 pKa = 3.14IRR316 pKa = 11.84IPRR319 pKa = 11.84FTFHH323 pKa = 7.65AFRR326 pKa = 11.84HH327 pKa = 4.69THH329 pKa = 6.92ASLLLNAGISYY340 pKa = 10.68KK341 pKa = 10.23EE342 pKa = 3.61LQYY345 pKa = 11.49RR346 pKa = 11.84LGHH349 pKa = 6.19ATLAMTMDD357 pKa = 4.24IYY359 pKa = 11.79SHH361 pKa = 6.95LSKK364 pKa = 10.92DD365 pKa = 3.51KK366 pKa = 10.41EE367 pKa = 4.25KK368 pKa = 10.93EE369 pKa = 3.85AVSYY373 pKa = 9.52YY374 pKa = 10.59EE375 pKa = 4.03KK376 pKa = 10.7AINGLL381 pKa = 3.67
Molecular weight: 43.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3359 |
41 |
490 |
197.6 |
22.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.639 ± 0.545 | 0.685 ± 0.196 |
5.061 ± 0.22 | 8.276 ± 0.511 |
4.376 ± 0.449 | 5.15 ± 0.334 |
1.786 ± 0.234 | 6.847 ± 0.421 |
8.247 ± 0.328 | 9.527 ± 0.458 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.292 ± 0.221 | 5.001 ± 0.658 |
3.96 ± 0.408 | 3.602 ± 0.484 |
5.567 ± 0.565 | 6.133 ± 0.493 |
6.103 ± 0.422 | 5.656 ± 0.584 |
0.774 ± 0.184 | 4.317 ± 0.325 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
