
CRESS virus sp. ctxk12
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; unclassified Cressdnaviricota; CRESS viruses
Average proteome isoelectric point is 7.8
Get precalculated fractions of proteins
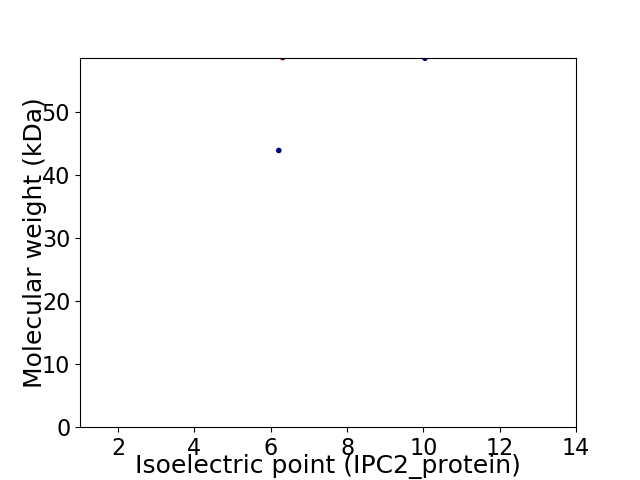
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
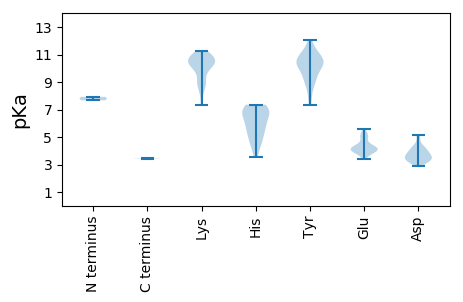
Protein with the lowest isoelectric point:
>tr|A0A5Q2W7X5|A0A5Q2W7X5_9VIRU Rep protein OS=CRESS virus sp. ctxk12 OX=2656692 PE=4 SV=1
MM1 pKa = 7.67PGRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84WTWTYY11 pKa = 11.27NNEE14 pKa = 4.33ALNHH18 pKa = 5.72ATTIEE23 pKa = 4.01QIEE26 pKa = 4.35WGDD29 pKa = 4.02KK30 pKa = 9.88IAADD34 pKa = 4.11LTRR37 pKa = 11.84VPDD40 pKa = 3.93MQWMFSLEE48 pKa = 4.02RR49 pKa = 11.84GKK51 pKa = 10.73SGNIHH56 pKa = 5.73LQGYY60 pKa = 8.98SKK62 pKa = 10.81LPRR65 pKa = 11.84AMSLAAMKK73 pKa = 10.15IVLPGAHH80 pKa = 6.98LLASRR85 pKa = 11.84GSTAQNKK92 pKa = 9.21AYY94 pKa = 10.17CGKK97 pKa = 10.24SDD99 pKa = 3.44THH101 pKa = 7.35IAGPWQSEE109 pKa = 4.06EE110 pKa = 4.94FIDD113 pKa = 5.12DD114 pKa = 3.86DD115 pKa = 4.22EE116 pKa = 4.29EE117 pKa = 5.19AKK119 pKa = 10.81EE120 pKa = 4.94KK121 pKa = 10.92EE122 pKa = 4.11DD123 pKa = 4.4LDD125 pKa = 4.15AEE127 pKa = 4.33KK128 pKa = 10.3EE129 pKa = 3.94AAAQAVIKK137 pKa = 10.31CAEE140 pKa = 4.31EE141 pKa = 3.89GHH143 pKa = 6.66LVQDD147 pKa = 3.58MFKK150 pKa = 10.71LLKK153 pKa = 10.33DD154 pKa = 3.07KK155 pKa = 10.03RR156 pKa = 11.84TRR158 pKa = 11.84GQTIHH163 pKa = 6.58MLPALRR169 pKa = 11.84LASTGLPPPIIARR182 pKa = 11.84GPLDD186 pKa = 3.46IEE188 pKa = 4.29LHH190 pKa = 6.31FGDD193 pKa = 4.1TGLGKK198 pKa = 8.66TFHH201 pKa = 6.19VRR203 pKa = 11.84RR204 pKa = 11.84KK205 pKa = 10.49YY206 pKa = 9.96PDD208 pKa = 3.47SYY210 pKa = 11.7LKK212 pKa = 10.67DD213 pKa = 3.55FHH215 pKa = 6.96GKK217 pKa = 9.03RR218 pKa = 11.84DD219 pKa = 3.42PWFDD223 pKa = 3.81GYY225 pKa = 11.09CGQTCLILDD234 pKa = 4.28DD235 pKa = 5.16FNGTGCPINTLKK247 pKa = 10.88RR248 pKa = 11.84LLDD251 pKa = 4.23CYY253 pKa = 10.49PCRR256 pKa = 11.84LQIKK260 pKa = 10.43GGTTWAAWTTAAITTQKK277 pKa = 11.09DD278 pKa = 3.18PIYY281 pKa = 9.25WYY283 pKa = 9.65PRR285 pKa = 11.84EE286 pKa = 4.38DD287 pKa = 3.5EE288 pKa = 4.52ADD290 pKa = 3.44RR291 pKa = 11.84KK292 pKa = 10.86AIFRR296 pKa = 11.84RR297 pKa = 11.84IARR300 pKa = 11.84VYY302 pKa = 9.13QWSADD307 pKa = 3.13GSIRR311 pKa = 11.84WRR313 pKa = 11.84PGLDD317 pKa = 3.14HH318 pKa = 7.13FEE320 pKa = 4.22HH321 pKa = 7.35RR322 pKa = 11.84GMAWTVVDD330 pKa = 4.44LGGPEE335 pKa = 3.85GGEE338 pKa = 3.8ARR340 pKa = 11.84RR341 pKa = 11.84DD342 pKa = 3.3EE343 pKa = 4.77DD344 pKa = 3.51SAEE347 pKa = 4.1FTSGQSDD354 pKa = 3.5GGSWSRR360 pKa = 11.84GGDD363 pKa = 3.21GGGGGGRR370 pKa = 11.84EE371 pKa = 3.95RR372 pKa = 11.84QRR374 pKa = 11.84GGIPTDD380 pKa = 3.18AGSRR384 pKa = 11.84RR385 pKa = 11.84GEE387 pKa = 4.04WTHH390 pKa = 6.72GPVV393 pKa = 3.45
MM1 pKa = 7.67PGRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84WTWTYY11 pKa = 11.27NNEE14 pKa = 4.33ALNHH18 pKa = 5.72ATTIEE23 pKa = 4.01QIEE26 pKa = 4.35WGDD29 pKa = 4.02KK30 pKa = 9.88IAADD34 pKa = 4.11LTRR37 pKa = 11.84VPDD40 pKa = 3.93MQWMFSLEE48 pKa = 4.02RR49 pKa = 11.84GKK51 pKa = 10.73SGNIHH56 pKa = 5.73LQGYY60 pKa = 8.98SKK62 pKa = 10.81LPRR65 pKa = 11.84AMSLAAMKK73 pKa = 10.15IVLPGAHH80 pKa = 6.98LLASRR85 pKa = 11.84GSTAQNKK92 pKa = 9.21AYY94 pKa = 10.17CGKK97 pKa = 10.24SDD99 pKa = 3.44THH101 pKa = 7.35IAGPWQSEE109 pKa = 4.06EE110 pKa = 4.94FIDD113 pKa = 5.12DD114 pKa = 3.86DD115 pKa = 4.22EE116 pKa = 4.29EE117 pKa = 5.19AKK119 pKa = 10.81EE120 pKa = 4.94KK121 pKa = 10.92EE122 pKa = 4.11DD123 pKa = 4.4LDD125 pKa = 4.15AEE127 pKa = 4.33KK128 pKa = 10.3EE129 pKa = 3.94AAAQAVIKK137 pKa = 10.31CAEE140 pKa = 4.31EE141 pKa = 3.89GHH143 pKa = 6.66LVQDD147 pKa = 3.58MFKK150 pKa = 10.71LLKK153 pKa = 10.33DD154 pKa = 3.07KK155 pKa = 10.03RR156 pKa = 11.84TRR158 pKa = 11.84GQTIHH163 pKa = 6.58MLPALRR169 pKa = 11.84LASTGLPPPIIARR182 pKa = 11.84GPLDD186 pKa = 3.46IEE188 pKa = 4.29LHH190 pKa = 6.31FGDD193 pKa = 4.1TGLGKK198 pKa = 8.66TFHH201 pKa = 6.19VRR203 pKa = 11.84RR204 pKa = 11.84KK205 pKa = 10.49YY206 pKa = 9.96PDD208 pKa = 3.47SYY210 pKa = 11.7LKK212 pKa = 10.67DD213 pKa = 3.55FHH215 pKa = 6.96GKK217 pKa = 9.03RR218 pKa = 11.84DD219 pKa = 3.42PWFDD223 pKa = 3.81GYY225 pKa = 11.09CGQTCLILDD234 pKa = 4.28DD235 pKa = 5.16FNGTGCPINTLKK247 pKa = 10.88RR248 pKa = 11.84LLDD251 pKa = 4.23CYY253 pKa = 10.49PCRR256 pKa = 11.84LQIKK260 pKa = 10.43GGTTWAAWTTAAITTQKK277 pKa = 11.09DD278 pKa = 3.18PIYY281 pKa = 9.25WYY283 pKa = 9.65PRR285 pKa = 11.84EE286 pKa = 4.38DD287 pKa = 3.5EE288 pKa = 4.52ADD290 pKa = 3.44RR291 pKa = 11.84KK292 pKa = 10.86AIFRR296 pKa = 11.84RR297 pKa = 11.84IARR300 pKa = 11.84VYY302 pKa = 9.13QWSADD307 pKa = 3.13GSIRR311 pKa = 11.84WRR313 pKa = 11.84PGLDD317 pKa = 3.14HH318 pKa = 7.13FEE320 pKa = 4.22HH321 pKa = 7.35RR322 pKa = 11.84GMAWTVVDD330 pKa = 4.44LGGPEE335 pKa = 3.85GGEE338 pKa = 3.8ARR340 pKa = 11.84RR341 pKa = 11.84DD342 pKa = 3.3EE343 pKa = 4.77DD344 pKa = 3.51SAEE347 pKa = 4.1FTSGQSDD354 pKa = 3.5GGSWSRR360 pKa = 11.84GGDD363 pKa = 3.21GGGGGGRR370 pKa = 11.84EE371 pKa = 3.95RR372 pKa = 11.84QRR374 pKa = 11.84GGIPTDD380 pKa = 3.18AGSRR384 pKa = 11.84RR385 pKa = 11.84GEE387 pKa = 4.04WTHH390 pKa = 6.72GPVV393 pKa = 3.45
Molecular weight: 43.87 kDa
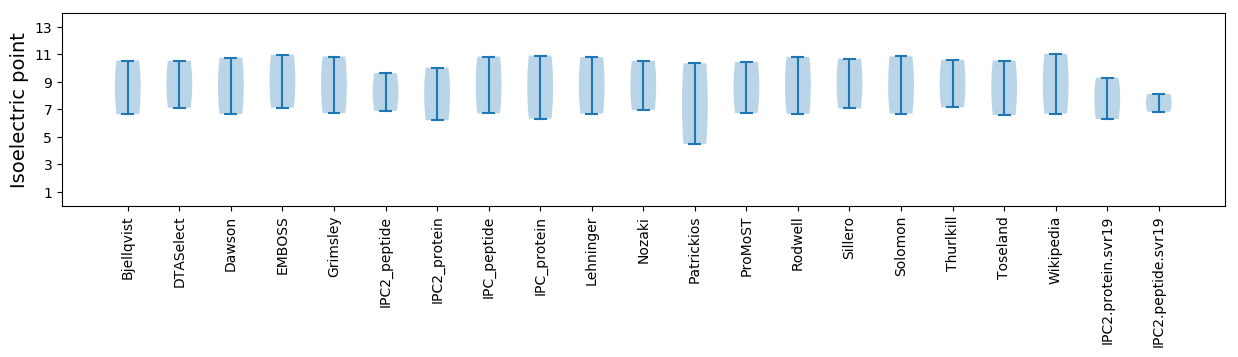
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W7X5|A0A5Q2W7X5_9VIRU Rep protein OS=CRESS virus sp. ctxk12 OX=2656692 PE=4 SV=1
MM1 pKa = 7.92PLPFFTAAGVAAAGIASYY19 pKa = 10.77LGIGTEE25 pKa = 4.13GRR27 pKa = 11.84EE28 pKa = 4.09ALGKK32 pKa = 10.78GLGQLSSAADD42 pKa = 3.62LALDD46 pKa = 3.73VAEE49 pKa = 5.57LYY51 pKa = 10.26PGAPWFSQGARR62 pKa = 11.84YY63 pKa = 9.33VRR65 pKa = 11.84DD66 pKa = 3.95YY67 pKa = 11.2ISGVHH72 pKa = 5.26GMHH75 pKa = 6.97RR76 pKa = 11.84KK77 pKa = 7.35VQGMLGDD84 pKa = 3.87RR85 pKa = 11.84DD86 pKa = 3.79VHH88 pKa = 5.75PVQVPGYY95 pKa = 7.3SEE97 pKa = 4.1RR98 pKa = 11.84VPYY101 pKa = 10.35DD102 pKa = 3.34SYY104 pKa = 12.07KK105 pKa = 10.28LDD107 pKa = 2.92SWGIPRR113 pKa = 11.84HH114 pKa = 3.54QTYY117 pKa = 8.29WVRR120 pKa = 11.84EE121 pKa = 4.09HH122 pKa = 7.17PSFFRR127 pKa = 11.84PYY129 pKa = 9.55VGRR132 pKa = 11.84RR133 pKa = 11.84SQLSYY138 pKa = 11.21NKK140 pKa = 8.72TRR142 pKa = 11.84APRR145 pKa = 11.84VGYY148 pKa = 8.54WRR150 pKa = 11.84PPLSAPRR157 pKa = 11.84RR158 pKa = 11.84SIMPRR163 pKa = 11.84RR164 pKa = 11.84FRR166 pKa = 11.84RR167 pKa = 11.84RR168 pKa = 11.84FTRR171 pKa = 11.84PRR173 pKa = 11.84FRR175 pKa = 11.84RR176 pKa = 11.84RR177 pKa = 11.84FRR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84YY182 pKa = 7.79GGYY185 pKa = 8.99RR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84RR189 pKa = 11.84SFGVKK194 pKa = 8.64RR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84FVYY200 pKa = 9.75RR201 pKa = 11.84RR202 pKa = 11.84ARR204 pKa = 11.84RR205 pKa = 11.84LIASMRR211 pKa = 11.84PKK213 pKa = 10.47RR214 pKa = 11.84ADD216 pKa = 3.09VQVGHH221 pKa = 6.96KK222 pKa = 10.16SVAANALTNQASPTFGASGNVLYY245 pKa = 10.14ISPMRR250 pKa = 11.84MPARR254 pKa = 11.84PGGQYY259 pKa = 10.68DD260 pKa = 3.07IGGTKK265 pKa = 10.34YY266 pKa = 10.52PLRR269 pKa = 11.84TAANQMQGRR278 pKa = 11.84KK279 pKa = 8.86IRR281 pKa = 11.84LHH283 pKa = 5.34RR284 pKa = 11.84LKK286 pKa = 10.79FKK288 pKa = 11.22FEE290 pKa = 3.66FWYY293 pKa = 10.09DD294 pKa = 3.39QSVMYY299 pKa = 10.7GLDD302 pKa = 3.55YY303 pKa = 11.35SGGAPTPAFSSFHH316 pKa = 4.51VWFGLFRR323 pKa = 11.84KK324 pKa = 10.27NIGSVANAYY333 pKa = 9.96AQFGLYY339 pKa = 9.85SDD341 pKa = 4.93SLNSSDD347 pKa = 4.66KK348 pKa = 10.63PYY350 pKa = 10.39IFYY353 pKa = 10.7RR354 pKa = 11.84RR355 pKa = 11.84NHH357 pKa = 6.34DD358 pKa = 3.67TLNTLGDD365 pKa = 3.9FSVGQQWHH373 pKa = 4.37QRR375 pKa = 11.84EE376 pKa = 3.77VRR378 pKa = 11.84YY379 pKa = 8.95WLRR382 pKa = 11.84HH383 pKa = 4.57PRR385 pKa = 11.84FLKK388 pKa = 9.82YY389 pKa = 10.36FKK391 pKa = 11.02LLGHH395 pKa = 5.53KK396 pKa = 9.45HH397 pKa = 4.88YY398 pKa = 10.71RR399 pKa = 11.84VGPEE403 pKa = 3.4QNIWARR409 pKa = 11.84TSAGTNVYY417 pKa = 10.29EE418 pKa = 5.15PIRR421 pKa = 11.84KK422 pKa = 9.08DD423 pKa = 3.3VAPVLGAAAGQFTFRR438 pKa = 11.84SRR440 pKa = 11.84KK441 pKa = 8.02GLVMEE446 pKa = 5.28RR447 pKa = 11.84DD448 pKa = 3.44QDD450 pKa = 4.29SVGVTPAALVDD461 pKa = 3.96GALPLYY467 pKa = 10.55AGQCDD472 pKa = 3.71AFQCVYY478 pKa = 10.67GAVPFMVIYY487 pKa = 10.97CEE489 pKa = 4.13DD490 pKa = 3.75TSGLTAEE497 pKa = 5.12ANWTLNSTYY506 pKa = 10.51AISWNEE512 pKa = 3.43
MM1 pKa = 7.92PLPFFTAAGVAAAGIASYY19 pKa = 10.77LGIGTEE25 pKa = 4.13GRR27 pKa = 11.84EE28 pKa = 4.09ALGKK32 pKa = 10.78GLGQLSSAADD42 pKa = 3.62LALDD46 pKa = 3.73VAEE49 pKa = 5.57LYY51 pKa = 10.26PGAPWFSQGARR62 pKa = 11.84YY63 pKa = 9.33VRR65 pKa = 11.84DD66 pKa = 3.95YY67 pKa = 11.2ISGVHH72 pKa = 5.26GMHH75 pKa = 6.97RR76 pKa = 11.84KK77 pKa = 7.35VQGMLGDD84 pKa = 3.87RR85 pKa = 11.84DD86 pKa = 3.79VHH88 pKa = 5.75PVQVPGYY95 pKa = 7.3SEE97 pKa = 4.1RR98 pKa = 11.84VPYY101 pKa = 10.35DD102 pKa = 3.34SYY104 pKa = 12.07KK105 pKa = 10.28LDD107 pKa = 2.92SWGIPRR113 pKa = 11.84HH114 pKa = 3.54QTYY117 pKa = 8.29WVRR120 pKa = 11.84EE121 pKa = 4.09HH122 pKa = 7.17PSFFRR127 pKa = 11.84PYY129 pKa = 9.55VGRR132 pKa = 11.84RR133 pKa = 11.84SQLSYY138 pKa = 11.21NKK140 pKa = 8.72TRR142 pKa = 11.84APRR145 pKa = 11.84VGYY148 pKa = 8.54WRR150 pKa = 11.84PPLSAPRR157 pKa = 11.84RR158 pKa = 11.84SIMPRR163 pKa = 11.84RR164 pKa = 11.84FRR166 pKa = 11.84RR167 pKa = 11.84RR168 pKa = 11.84FTRR171 pKa = 11.84PRR173 pKa = 11.84FRR175 pKa = 11.84RR176 pKa = 11.84RR177 pKa = 11.84FRR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84YY182 pKa = 7.79GGYY185 pKa = 8.99RR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84RR189 pKa = 11.84SFGVKK194 pKa = 8.64RR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84FVYY200 pKa = 9.75RR201 pKa = 11.84RR202 pKa = 11.84ARR204 pKa = 11.84RR205 pKa = 11.84LIASMRR211 pKa = 11.84PKK213 pKa = 10.47RR214 pKa = 11.84ADD216 pKa = 3.09VQVGHH221 pKa = 6.96KK222 pKa = 10.16SVAANALTNQASPTFGASGNVLYY245 pKa = 10.14ISPMRR250 pKa = 11.84MPARR254 pKa = 11.84PGGQYY259 pKa = 10.68DD260 pKa = 3.07IGGTKK265 pKa = 10.34YY266 pKa = 10.52PLRR269 pKa = 11.84TAANQMQGRR278 pKa = 11.84KK279 pKa = 8.86IRR281 pKa = 11.84LHH283 pKa = 5.34RR284 pKa = 11.84LKK286 pKa = 10.79FKK288 pKa = 11.22FEE290 pKa = 3.66FWYY293 pKa = 10.09DD294 pKa = 3.39QSVMYY299 pKa = 10.7GLDD302 pKa = 3.55YY303 pKa = 11.35SGGAPTPAFSSFHH316 pKa = 4.51VWFGLFRR323 pKa = 11.84KK324 pKa = 10.27NIGSVANAYY333 pKa = 9.96AQFGLYY339 pKa = 9.85SDD341 pKa = 4.93SLNSSDD347 pKa = 4.66KK348 pKa = 10.63PYY350 pKa = 10.39IFYY353 pKa = 10.7RR354 pKa = 11.84RR355 pKa = 11.84NHH357 pKa = 6.34DD358 pKa = 3.67TLNTLGDD365 pKa = 3.9FSVGQQWHH373 pKa = 4.37QRR375 pKa = 11.84EE376 pKa = 3.77VRR378 pKa = 11.84YY379 pKa = 8.95WLRR382 pKa = 11.84HH383 pKa = 4.57PRR385 pKa = 11.84FLKK388 pKa = 9.82YY389 pKa = 10.36FKK391 pKa = 11.02LLGHH395 pKa = 5.53KK396 pKa = 9.45HH397 pKa = 4.88YY398 pKa = 10.71RR399 pKa = 11.84VGPEE403 pKa = 3.4QNIWARR409 pKa = 11.84TSAGTNVYY417 pKa = 10.29EE418 pKa = 5.15PIRR421 pKa = 11.84KK422 pKa = 9.08DD423 pKa = 3.3VAPVLGAAAGQFTFRR438 pKa = 11.84SRR440 pKa = 11.84KK441 pKa = 8.02GLVMEE446 pKa = 5.28RR447 pKa = 11.84DD448 pKa = 3.44QDD450 pKa = 4.29SVGVTPAALVDD461 pKa = 3.96GALPLYY467 pKa = 10.55AGQCDD472 pKa = 3.71AFQCVYY478 pKa = 10.67GAVPFMVIYY487 pKa = 10.97CEE489 pKa = 4.13DD490 pKa = 3.75TSGLTAEE497 pKa = 5.12ANWTLNSTYY506 pKa = 10.51AISWNEE512 pKa = 3.43
Molecular weight: 58.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
905 |
393 |
512 |
452.5 |
51.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.84 ± 0.12 | 1.105 ± 0.431 |
5.746 ± 1.366 | 4.088 ± 1.287 |
4.309 ± 0.963 | 10.387 ± 0.841 |
2.762 ± 0.186 | 3.978 ± 0.871 |
4.309 ± 0.66 | 7.072 ± 0.196 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.099 ± 0.041 | 2.431 ± 0.414 |
5.746 ± 0.419 | 3.867 ± 0.195 |
10.276 ± 1.361 | 5.856 ± 0.814 |
4.972 ± 0.886 | 4.751 ± 1.57 |
2.762 ± 0.51 | 4.641 ± 1.337 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
