
Torque teno virus 21
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 7.67
Get precalculated fractions of proteins
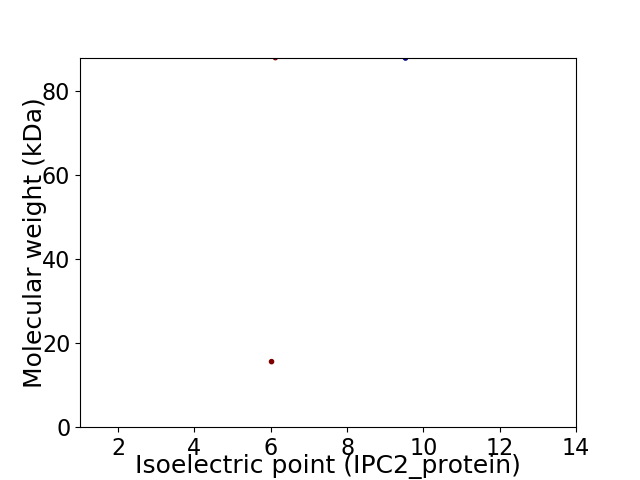
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q98Y63|Q98Y63_9VIRU Capsid protein OS=Torque teno virus 21 OX=687360 PE=3 SV=1
MM1 pKa = 7.68RR2 pKa = 11.84FSRR5 pKa = 11.84IYY7 pKa = 9.92RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.97KK11 pKa = 10.12RR12 pKa = 11.84LLPLLLVPTEE22 pKa = 4.1PKK24 pKa = 10.14EE25 pKa = 3.85QFVMSWRR32 pKa = 11.84CPLEE36 pKa = 3.76NAYY39 pKa = 9.68KK40 pKa = 10.56RR41 pKa = 11.84EE42 pKa = 3.99INFLRR47 pKa = 11.84GCQMLHH53 pKa = 5.64TCFCGCDD60 pKa = 3.39DD61 pKa = 5.84FINHH65 pKa = 7.84IIRR68 pKa = 11.84LQNLHH73 pKa = 6.74GNLHH77 pKa = 5.89QPTGPSTPPVGRR89 pKa = 11.84RR90 pKa = 11.84ALALPAAPEE99 pKa = 3.96PWRR102 pKa = 11.84GDD104 pKa = 3.14GGGPEE109 pKa = 4.52GDD111 pKa = 3.32RR112 pKa = 11.84TADD115 pKa = 3.44GPADD119 pKa = 3.73AGGDD123 pKa = 3.93YY124 pKa = 11.22APGDD128 pKa = 4.07LDD130 pKa = 5.27DD131 pKa = 5.65LFAAAAADD139 pKa = 3.75QEE141 pKa = 4.47
MM1 pKa = 7.68RR2 pKa = 11.84FSRR5 pKa = 11.84IYY7 pKa = 9.92RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.97KK11 pKa = 10.12RR12 pKa = 11.84LLPLLLVPTEE22 pKa = 4.1PKK24 pKa = 10.14EE25 pKa = 3.85QFVMSWRR32 pKa = 11.84CPLEE36 pKa = 3.76NAYY39 pKa = 9.68KK40 pKa = 10.56RR41 pKa = 11.84EE42 pKa = 3.99INFLRR47 pKa = 11.84GCQMLHH53 pKa = 5.64TCFCGCDD60 pKa = 3.39DD61 pKa = 5.84FINHH65 pKa = 7.84IIRR68 pKa = 11.84LQNLHH73 pKa = 6.74GNLHH77 pKa = 5.89QPTGPSTPPVGRR89 pKa = 11.84RR90 pKa = 11.84ALALPAAPEE99 pKa = 3.96PWRR102 pKa = 11.84GDD104 pKa = 3.14GGGPEE109 pKa = 4.52GDD111 pKa = 3.32RR112 pKa = 11.84TADD115 pKa = 3.44GPADD119 pKa = 3.73AGGDD123 pKa = 3.93YY124 pKa = 11.22APGDD128 pKa = 4.07LDD130 pKa = 5.27DD131 pKa = 5.65LFAAAAADD139 pKa = 3.75QEE141 pKa = 4.47
Molecular weight: 15.63 kDa
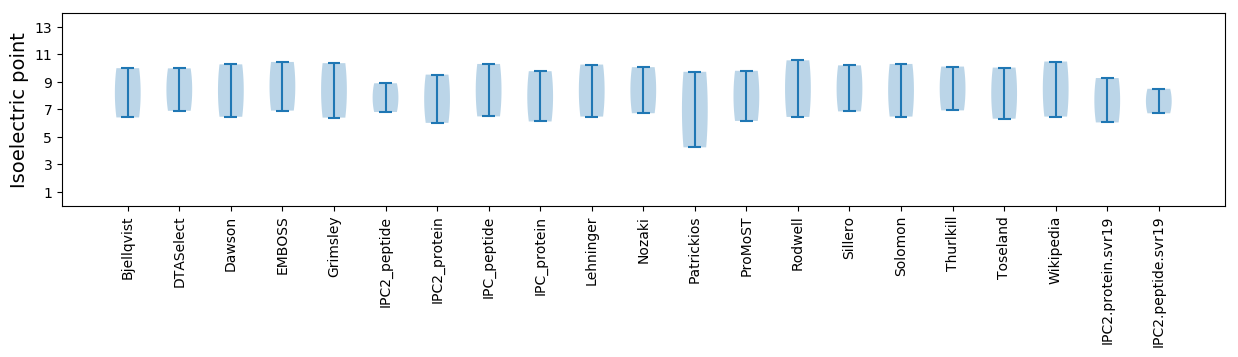
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q98Y63|Q98Y63_9VIRU Capsid protein OS=Torque teno virus 21 OX=687360 PE=3 SV=1
MM1 pKa = 7.59AWRR4 pKa = 11.84WWWARR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84PNRR14 pKa = 11.84RR15 pKa = 11.84WTRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84WRR22 pKa = 11.84RR23 pKa = 11.84LRR25 pKa = 11.84TRR27 pKa = 11.84RR28 pKa = 11.84PRR30 pKa = 11.84RR31 pKa = 11.84PVRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84PRR41 pKa = 11.84VRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84WGRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84GRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84FYY57 pKa = 10.38RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84FKK62 pKa = 10.46KK63 pKa = 9.34RR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84KK70 pKa = 8.68LTLKK74 pKa = 8.77MWNPSTIRR82 pKa = 11.84ACNIKK87 pKa = 10.5GAINLVMCGHH97 pKa = 6.2TQAGRR102 pKa = 11.84NYY104 pKa = 10.21AIRR107 pKa = 11.84SEE109 pKa = 4.31DD110 pKa = 4.29FYY112 pKa = 11.18PQIQSFGGSFSTTTWSLRR130 pKa = 11.84VLFDD134 pKa = 4.0EE135 pKa = 4.33YY136 pKa = 11.46QKK138 pKa = 11.11FHH140 pKa = 6.8NFWTYY145 pKa = 11.35PNTQLDD151 pKa = 3.88LCRR154 pKa = 11.84YY155 pKa = 8.82KK156 pKa = 10.82YY157 pKa = 10.79AIFTFYY163 pKa = 10.72RR164 pKa = 11.84DD165 pKa = 3.52PKK167 pKa = 9.57VDD169 pKa = 3.78YY170 pKa = 10.75IVIYY174 pKa = 8.74NTNPPFKK181 pKa = 9.61INKK184 pKa = 7.99YY185 pKa = 9.82SSPFLHH191 pKa = 7.21PGLMMLQKK199 pKa = 10.48KK200 pKa = 9.99KK201 pKa = 10.62ILIPSFQTKK210 pKa = 9.88PGGKK214 pKa = 9.32SRR216 pKa = 11.84IKK218 pKa = 11.0VKK220 pKa = 10.42IKK222 pKa = 10.4PPALFEE228 pKa = 4.91DD229 pKa = 3.32KK230 pKa = 10.27WYY232 pKa = 8.51TQQDD236 pKa = 4.1LCPVNLLSLAVSACSFIHH254 pKa = 6.19PFCSPEE260 pKa = 3.81SDD262 pKa = 3.99TICMTFQVLRR272 pKa = 11.84EE273 pKa = 4.09FYY275 pKa = 8.51YY276 pKa = 9.99THH278 pKa = 6.71LTVTPTTTTSTPEE291 pKa = 3.61KK292 pKa = 10.18DD293 pKa = 3.12KK294 pKa = 11.42KK295 pKa = 10.73IFNDD299 pKa = 3.46QLYY302 pKa = 10.67SNANFYY308 pKa = 10.97QSLHH312 pKa = 6.25ASAFLNIAQAPAIHH326 pKa = 6.15GHH328 pKa = 5.99NGIPNNSRR336 pKa = 11.84YY337 pKa = 9.82LSSTGTEE344 pKa = 3.91TSFRR348 pKa = 11.84TGNNSIYY355 pKa = 10.31GQPNYY360 pKa = 10.72KK361 pKa = 10.03PIPEE365 pKa = 4.5KK366 pKa = 10.17LTEE369 pKa = 3.57IRR371 pKa = 11.84KK372 pKa = 8.88WFFKK376 pKa = 10.65QATTPNEE383 pKa = 3.51IHH385 pKa = 5.89GTYY388 pKa = 10.53GKK390 pKa = 8.22PTYY393 pKa = 10.36DD394 pKa = 3.12AVDD397 pKa = 3.58YY398 pKa = 11.06HH399 pKa = 7.18LGKK402 pKa = 10.59YY403 pKa = 10.29SPIFLSPYY411 pKa = 8.3RR412 pKa = 11.84TNTQFPTAYY421 pKa = 9.54MDD423 pKa = 3.17VTYY426 pKa = 11.01NPNVDD431 pKa = 3.13KK432 pKa = 11.55GKK434 pKa = 11.02GNKK437 pKa = 8.61IWLQSVTKK445 pKa = 8.64EE446 pKa = 3.97TSDD449 pKa = 3.47FDD451 pKa = 4.32SRR453 pKa = 11.84SCRR456 pKa = 11.84CIIEE460 pKa = 4.21NLPMWAMVNGYY471 pKa = 10.42SDD473 pKa = 3.64FAEE476 pKa = 4.33SEE478 pKa = 4.31LGSEE482 pKa = 3.9VHH484 pKa = 6.74AVYY487 pKa = 10.17VCCIICPYY495 pKa = 8.42TKK497 pKa = 10.48PMLYY501 pKa = 10.65NKK503 pKa = 7.9TNPAMGYY510 pKa = 9.53IFYY513 pKa = 8.09DD514 pKa = 3.56TLFGDD519 pKa = 4.68GKK521 pKa = 10.67LPSGPGLVPFYY532 pKa = 9.76WQSRR536 pKa = 11.84WYY538 pKa = 9.56PKK540 pKa = 10.26LAWQQQVLHH549 pKa = 7.02DD550 pKa = 5.02FYY552 pKa = 11.68LCGPFSYY559 pKa = 10.95KK560 pKa = 10.47DD561 pKa = 3.51DD562 pKa = 4.29LKK564 pKa = 11.41SFTINTTYY572 pKa = 10.88KK573 pKa = 10.66FKK575 pKa = 10.82FLWGGNMIPEE585 pKa = 3.97QVIKK589 pKa = 10.67NPCKK593 pKa = 8.69TTDD596 pKa = 3.33PTYY599 pKa = 10.33TLSDD603 pKa = 3.43RR604 pKa = 11.84QRR606 pKa = 11.84RR607 pKa = 11.84DD608 pKa = 3.36LQVVDD613 pKa = 5.68PITMGPQWEE622 pKa = 4.23FHH624 pKa = 4.97TWDD627 pKa = 2.9WRR629 pKa = 11.84RR630 pKa = 11.84GLFGQNALRR639 pKa = 11.84RR640 pKa = 11.84VSEE643 pKa = 4.14KK644 pKa = 10.77PGDD647 pKa = 3.77DD648 pKa = 3.53AEE650 pKa = 4.73YY651 pKa = 10.24YY652 pKa = 10.82APPKK656 pKa = 10.03KK657 pKa = 9.93PRR659 pKa = 11.84FFPPTDD665 pKa = 3.52LEE667 pKa = 4.23EE668 pKa = 4.08QEE670 pKa = 4.84KK671 pKa = 11.16DD672 pKa = 3.13SDD674 pKa = 4.11SQEE677 pKa = 3.81EE678 pKa = 4.32TRR680 pKa = 11.84LLFHH684 pKa = 7.03PSPPRR689 pKa = 11.84SQEE692 pKa = 3.9EE693 pKa = 4.0IQQEE697 pKa = 4.18QQRR700 pKa = 11.84DD701 pKa = 2.77IHH703 pKa = 6.9LRR705 pKa = 11.84LGQQLRR711 pKa = 11.84IRR713 pKa = 11.84QQLQQVFLQVLKK725 pKa = 9.24TQANLHH731 pKa = 6.13INPLFLNQQQ740 pKa = 3.11
MM1 pKa = 7.59AWRR4 pKa = 11.84WWWARR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84PNRR14 pKa = 11.84RR15 pKa = 11.84WTRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84WRR22 pKa = 11.84RR23 pKa = 11.84LRR25 pKa = 11.84TRR27 pKa = 11.84RR28 pKa = 11.84PRR30 pKa = 11.84RR31 pKa = 11.84PVRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84PRR41 pKa = 11.84VRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84WGRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84GRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84FYY57 pKa = 10.38RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84FKK62 pKa = 10.46KK63 pKa = 9.34RR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84KK70 pKa = 8.68LTLKK74 pKa = 8.77MWNPSTIRR82 pKa = 11.84ACNIKK87 pKa = 10.5GAINLVMCGHH97 pKa = 6.2TQAGRR102 pKa = 11.84NYY104 pKa = 10.21AIRR107 pKa = 11.84SEE109 pKa = 4.31DD110 pKa = 4.29FYY112 pKa = 11.18PQIQSFGGSFSTTTWSLRR130 pKa = 11.84VLFDD134 pKa = 4.0EE135 pKa = 4.33YY136 pKa = 11.46QKK138 pKa = 11.11FHH140 pKa = 6.8NFWTYY145 pKa = 11.35PNTQLDD151 pKa = 3.88LCRR154 pKa = 11.84YY155 pKa = 8.82KK156 pKa = 10.82YY157 pKa = 10.79AIFTFYY163 pKa = 10.72RR164 pKa = 11.84DD165 pKa = 3.52PKK167 pKa = 9.57VDD169 pKa = 3.78YY170 pKa = 10.75IVIYY174 pKa = 8.74NTNPPFKK181 pKa = 9.61INKK184 pKa = 7.99YY185 pKa = 9.82SSPFLHH191 pKa = 7.21PGLMMLQKK199 pKa = 10.48KK200 pKa = 9.99KK201 pKa = 10.62ILIPSFQTKK210 pKa = 9.88PGGKK214 pKa = 9.32SRR216 pKa = 11.84IKK218 pKa = 11.0VKK220 pKa = 10.42IKK222 pKa = 10.4PPALFEE228 pKa = 4.91DD229 pKa = 3.32KK230 pKa = 10.27WYY232 pKa = 8.51TQQDD236 pKa = 4.1LCPVNLLSLAVSACSFIHH254 pKa = 6.19PFCSPEE260 pKa = 3.81SDD262 pKa = 3.99TICMTFQVLRR272 pKa = 11.84EE273 pKa = 4.09FYY275 pKa = 8.51YY276 pKa = 9.99THH278 pKa = 6.71LTVTPTTTTSTPEE291 pKa = 3.61KK292 pKa = 10.18DD293 pKa = 3.12KK294 pKa = 11.42KK295 pKa = 10.73IFNDD299 pKa = 3.46QLYY302 pKa = 10.67SNANFYY308 pKa = 10.97QSLHH312 pKa = 6.25ASAFLNIAQAPAIHH326 pKa = 6.15GHH328 pKa = 5.99NGIPNNSRR336 pKa = 11.84YY337 pKa = 9.82LSSTGTEE344 pKa = 3.91TSFRR348 pKa = 11.84TGNNSIYY355 pKa = 10.31GQPNYY360 pKa = 10.72KK361 pKa = 10.03PIPEE365 pKa = 4.5KK366 pKa = 10.17LTEE369 pKa = 3.57IRR371 pKa = 11.84KK372 pKa = 8.88WFFKK376 pKa = 10.65QATTPNEE383 pKa = 3.51IHH385 pKa = 5.89GTYY388 pKa = 10.53GKK390 pKa = 8.22PTYY393 pKa = 10.36DD394 pKa = 3.12AVDD397 pKa = 3.58YY398 pKa = 11.06HH399 pKa = 7.18LGKK402 pKa = 10.59YY403 pKa = 10.29SPIFLSPYY411 pKa = 8.3RR412 pKa = 11.84TNTQFPTAYY421 pKa = 9.54MDD423 pKa = 3.17VTYY426 pKa = 11.01NPNVDD431 pKa = 3.13KK432 pKa = 11.55GKK434 pKa = 11.02GNKK437 pKa = 8.61IWLQSVTKK445 pKa = 8.64EE446 pKa = 3.97TSDD449 pKa = 3.47FDD451 pKa = 4.32SRR453 pKa = 11.84SCRR456 pKa = 11.84CIIEE460 pKa = 4.21NLPMWAMVNGYY471 pKa = 10.42SDD473 pKa = 3.64FAEE476 pKa = 4.33SEE478 pKa = 4.31LGSEE482 pKa = 3.9VHH484 pKa = 6.74AVYY487 pKa = 10.17VCCIICPYY495 pKa = 8.42TKK497 pKa = 10.48PMLYY501 pKa = 10.65NKK503 pKa = 7.9TNPAMGYY510 pKa = 9.53IFYY513 pKa = 8.09DD514 pKa = 3.56TLFGDD519 pKa = 4.68GKK521 pKa = 10.67LPSGPGLVPFYY532 pKa = 9.76WQSRR536 pKa = 11.84WYY538 pKa = 9.56PKK540 pKa = 10.26LAWQQQVLHH549 pKa = 7.02DD550 pKa = 5.02FYY552 pKa = 11.68LCGPFSYY559 pKa = 10.95KK560 pKa = 10.47DD561 pKa = 3.51DD562 pKa = 4.29LKK564 pKa = 11.41SFTINTTYY572 pKa = 10.88KK573 pKa = 10.66FKK575 pKa = 10.82FLWGGNMIPEE585 pKa = 3.97QVIKK589 pKa = 10.67NPCKK593 pKa = 8.69TTDD596 pKa = 3.33PTYY599 pKa = 10.33TLSDD603 pKa = 3.43RR604 pKa = 11.84QRR606 pKa = 11.84RR607 pKa = 11.84DD608 pKa = 3.36LQVVDD613 pKa = 5.68PITMGPQWEE622 pKa = 4.23FHH624 pKa = 4.97TWDD627 pKa = 2.9WRR629 pKa = 11.84RR630 pKa = 11.84GLFGQNALRR639 pKa = 11.84RR640 pKa = 11.84VSEE643 pKa = 4.14KK644 pKa = 10.77PGDD647 pKa = 3.77DD648 pKa = 3.53AEE650 pKa = 4.73YY651 pKa = 10.24YY652 pKa = 10.82APPKK656 pKa = 10.03KK657 pKa = 9.93PRR659 pKa = 11.84FFPPTDD665 pKa = 3.52LEE667 pKa = 4.23EE668 pKa = 4.08QEE670 pKa = 4.84KK671 pKa = 11.16DD672 pKa = 3.13SDD674 pKa = 4.11SQEE677 pKa = 3.81EE678 pKa = 4.32TRR680 pKa = 11.84LLFHH684 pKa = 7.03PSPPRR689 pKa = 11.84SQEE692 pKa = 3.9EE693 pKa = 4.0IQQEE697 pKa = 4.18QQRR700 pKa = 11.84DD701 pKa = 2.77IHH703 pKa = 6.9LRR705 pKa = 11.84LGQQLRR711 pKa = 11.84IRR713 pKa = 11.84QQLQQVFLQVLKK725 pKa = 9.24TQANLHH731 pKa = 6.13INPLFLNQQQ740 pKa = 3.11
Molecular weight: 87.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
881 |
141 |
740 |
440.5 |
51.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.767 ± 2.398 | 2.157 ± 0.646 |
4.994 ± 1.304 | 3.859 ± 0.514 |
5.562 ± 0.607 | 5.562 ± 2.029 |
2.27 ± 0.263 | 4.881 ± 0.62 |
5.789 ± 1.371 | 7.832 ± 1.304 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.816 ± 0.145 | 4.767 ± 0.567 |
7.946 ± 0.922 | 5.448 ± 0.884 |
9.535 ± 0.146 | 5.221 ± 1.437 |
6.697 ± 1.464 | 3.405 ± 0.594 |
2.611 ± 0.554 | 4.881 ± 1.279 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
