
Tortoise microvirus 39
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.66
Get precalculated fractions of proteins
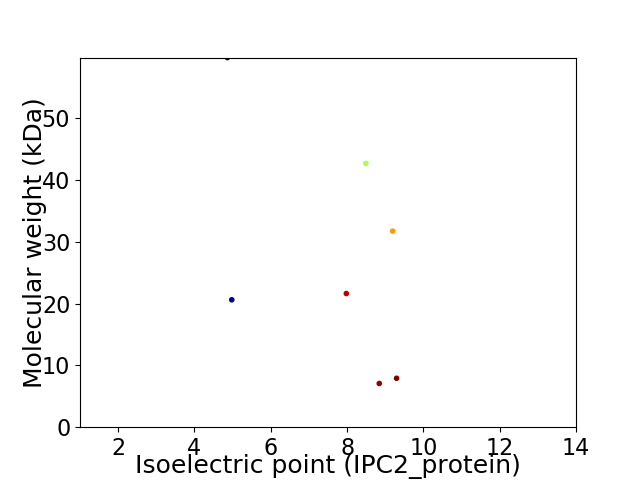
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W6F9|A0A4P8W6F9_9VIRU Uncharacterized protein OS=Tortoise microvirus 39 OX=2583142 PE=4 SV=1
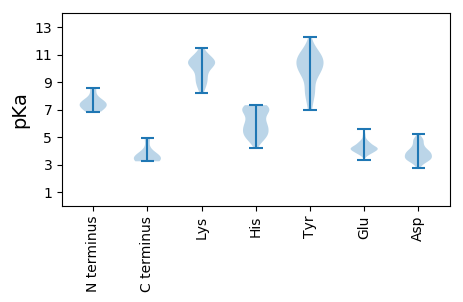
MM1 pKa = 7.21RR2 pKa = 11.84TSPVPVQRR10 pKa = 11.84TMRR13 pKa = 11.84RR14 pKa = 11.84HH15 pKa = 5.58DD16 pKa = 4.11LKK18 pKa = 11.47VLTSLPAGKK27 pKa = 9.52FVPLAAVPLLRR38 pKa = 11.84EE39 pKa = 4.22DD40 pKa = 3.63AVTRR44 pKa = 11.84LNGTISFEE52 pKa = 3.85MSEE55 pKa = 4.26TVEE58 pKa = 3.79ILMNAVNVRR67 pKa = 11.84VMAYY71 pKa = 9.58LVPFLAFEE79 pKa = 4.64RR80 pKa = 11.84FAGMDD85 pKa = 3.76DD86 pKa = 4.68LNRR89 pKa = 11.84SYY91 pKa = 11.4QKK93 pKa = 11.03VPMKK97 pKa = 10.79DD98 pKa = 3.39GDD100 pKa = 3.82PVIPFFEE107 pKa = 4.78SMVAPAHH114 pKa = 5.66GANPILTYY122 pKa = 9.63MGKK125 pKa = 8.94HH126 pKa = 5.81CRR128 pKa = 11.84PGTNINSAYY137 pKa = 9.63IEE139 pKa = 4.61AYY141 pKa = 8.58NAVWNFRR148 pKa = 11.84AKK150 pKa = 10.38NRR152 pKa = 11.84SPNITQRR159 pKa = 11.84TRR161 pKa = 11.84LEE163 pKa = 3.92ATLAPAFWLHH173 pKa = 6.62SNFNHH178 pKa = 5.72VVPDD182 pKa = 3.91FDD184 pKa = 3.59QAVIDD189 pKa = 4.87GEE191 pKa = 4.48VALNSAQVNLALKK204 pKa = 10.19NADD207 pKa = 3.86GTPGSAPVSVTGASLTVNSATSSTRR232 pKa = 11.84AVQMNNTGQLVYY244 pKa = 10.72AGSPLAAAAGVTGLGGLTADD264 pKa = 4.85LAAITAEE271 pKa = 4.07LQQDD275 pKa = 4.91GITISLSNIEE285 pKa = 4.05LARR288 pKa = 11.84RR289 pKa = 11.84TQAFAKK295 pKa = 10.31LRR297 pKa = 11.84AQYY300 pKa = 9.85HH301 pKa = 4.72GHH303 pKa = 5.68SDD305 pKa = 3.46EE306 pKa = 4.76YY307 pKa = 10.8IIDD310 pKa = 4.05LLMSGISVPEE320 pKa = 4.03QAWKK324 pKa = 10.57QPILLADD331 pKa = 3.37EE332 pKa = 4.24HH333 pKa = 6.95TIFGMSKK340 pKa = 10.01RR341 pKa = 11.84YY342 pKa = 10.09ASDD345 pKa = 3.21AANLTEE351 pKa = 4.22SVVNGGTVIDD361 pKa = 4.8LSVSVPRR368 pKa = 11.84VPTGGVVMLFAEE380 pKa = 4.62ITPEE384 pKa = 3.85QLFEE388 pKa = 4.21RR389 pKa = 11.84MKK391 pKa = 11.25DD392 pKa = 3.66PFLHH396 pKa = 7.02ALDD399 pKa = 4.83PQDD402 pKa = 4.94DD403 pKa = 4.06LPEE406 pKa = 4.27FVRR409 pKa = 11.84DD410 pKa = 3.48TLDD413 pKa = 3.51PEE415 pKa = 4.37KK416 pKa = 11.04VSVVPCEE423 pKa = 4.15YY424 pKa = 11.11VDD426 pKa = 4.11MDD428 pKa = 4.69HH429 pKa = 7.26DD430 pKa = 4.49TPDD433 pKa = 3.27STFGYY438 pKa = 10.89APLNYY443 pKa = 9.41EE444 pKa = 3.95WAQNSPQIGGRR455 pKa = 11.84FYY457 pKa = 11.2RR458 pKa = 11.84PAVDD462 pKa = 3.43AAFDD466 pKa = 3.88EE467 pKa = 4.45DD468 pKa = 3.97RR469 pKa = 11.84QRR471 pKa = 11.84IWAVEE476 pKa = 4.16TQNPTLSEE484 pKa = 3.99DD485 pKa = 4.19FYY487 pKa = 11.6LCTTMHH493 pKa = 6.09QKK495 pKa = 10.5PFVVTNVDD503 pKa = 3.2PFEE506 pKa = 5.02ALLRR510 pKa = 11.84GEE512 pKa = 5.11AIISGNTVFGGLLVEE527 pKa = 5.22ANDD530 pKa = 4.76DD531 pKa = 3.98YY532 pKa = 12.24AEE534 pKa = 4.31VAAKK538 pKa = 10.64APVDD542 pKa = 4.49RR543 pKa = 11.84IVKK546 pKa = 9.08PAA548 pKa = 3.47
MM1 pKa = 7.21RR2 pKa = 11.84TSPVPVQRR10 pKa = 11.84TMRR13 pKa = 11.84RR14 pKa = 11.84HH15 pKa = 5.58DD16 pKa = 4.11LKK18 pKa = 11.47VLTSLPAGKK27 pKa = 9.52FVPLAAVPLLRR38 pKa = 11.84EE39 pKa = 4.22DD40 pKa = 3.63AVTRR44 pKa = 11.84LNGTISFEE52 pKa = 3.85MSEE55 pKa = 4.26TVEE58 pKa = 3.79ILMNAVNVRR67 pKa = 11.84VMAYY71 pKa = 9.58LVPFLAFEE79 pKa = 4.64RR80 pKa = 11.84FAGMDD85 pKa = 3.76DD86 pKa = 4.68LNRR89 pKa = 11.84SYY91 pKa = 11.4QKK93 pKa = 11.03VPMKK97 pKa = 10.79DD98 pKa = 3.39GDD100 pKa = 3.82PVIPFFEE107 pKa = 4.78SMVAPAHH114 pKa = 5.66GANPILTYY122 pKa = 9.63MGKK125 pKa = 8.94HH126 pKa = 5.81CRR128 pKa = 11.84PGTNINSAYY137 pKa = 9.63IEE139 pKa = 4.61AYY141 pKa = 8.58NAVWNFRR148 pKa = 11.84AKK150 pKa = 10.38NRR152 pKa = 11.84SPNITQRR159 pKa = 11.84TRR161 pKa = 11.84LEE163 pKa = 3.92ATLAPAFWLHH173 pKa = 6.62SNFNHH178 pKa = 5.72VVPDD182 pKa = 3.91FDD184 pKa = 3.59QAVIDD189 pKa = 4.87GEE191 pKa = 4.48VALNSAQVNLALKK204 pKa = 10.19NADD207 pKa = 3.86GTPGSAPVSVTGASLTVNSATSSTRR232 pKa = 11.84AVQMNNTGQLVYY244 pKa = 10.72AGSPLAAAAGVTGLGGLTADD264 pKa = 4.85LAAITAEE271 pKa = 4.07LQQDD275 pKa = 4.91GITISLSNIEE285 pKa = 4.05LARR288 pKa = 11.84RR289 pKa = 11.84TQAFAKK295 pKa = 10.31LRR297 pKa = 11.84AQYY300 pKa = 9.85HH301 pKa = 4.72GHH303 pKa = 5.68SDD305 pKa = 3.46EE306 pKa = 4.76YY307 pKa = 10.8IIDD310 pKa = 4.05LLMSGISVPEE320 pKa = 4.03QAWKK324 pKa = 10.57QPILLADD331 pKa = 3.37EE332 pKa = 4.24HH333 pKa = 6.95TIFGMSKK340 pKa = 10.01RR341 pKa = 11.84YY342 pKa = 10.09ASDD345 pKa = 3.21AANLTEE351 pKa = 4.22SVVNGGTVIDD361 pKa = 4.8LSVSVPRR368 pKa = 11.84VPTGGVVMLFAEE380 pKa = 4.62ITPEE384 pKa = 3.85QLFEE388 pKa = 4.21RR389 pKa = 11.84MKK391 pKa = 11.25DD392 pKa = 3.66PFLHH396 pKa = 7.02ALDD399 pKa = 4.83PQDD402 pKa = 4.94DD403 pKa = 4.06LPEE406 pKa = 4.27FVRR409 pKa = 11.84DD410 pKa = 3.48TLDD413 pKa = 3.51PEE415 pKa = 4.37KK416 pKa = 11.04VSVVPCEE423 pKa = 4.15YY424 pKa = 11.11VDD426 pKa = 4.11MDD428 pKa = 4.69HH429 pKa = 7.26DD430 pKa = 4.49TPDD433 pKa = 3.27STFGYY438 pKa = 10.89APLNYY443 pKa = 9.41EE444 pKa = 3.95WAQNSPQIGGRR455 pKa = 11.84FYY457 pKa = 11.2RR458 pKa = 11.84PAVDD462 pKa = 3.43AAFDD466 pKa = 3.88EE467 pKa = 4.45DD468 pKa = 3.97RR469 pKa = 11.84QRR471 pKa = 11.84IWAVEE476 pKa = 4.16TQNPTLSEE484 pKa = 3.99DD485 pKa = 4.19FYY487 pKa = 11.6LCTTMHH493 pKa = 6.09QKK495 pKa = 10.5PFVVTNVDD503 pKa = 3.2PFEE506 pKa = 5.02ALLRR510 pKa = 11.84GEE512 pKa = 5.11AIISGNTVFGGLLVEE527 pKa = 5.22ANDD530 pKa = 4.76DD531 pKa = 3.98YY532 pKa = 12.24AEE534 pKa = 4.31VAAKK538 pKa = 10.64APVDD542 pKa = 4.49RR543 pKa = 11.84IVKK546 pKa = 9.08PAA548 pKa = 3.47
Molecular weight: 59.84 kDa
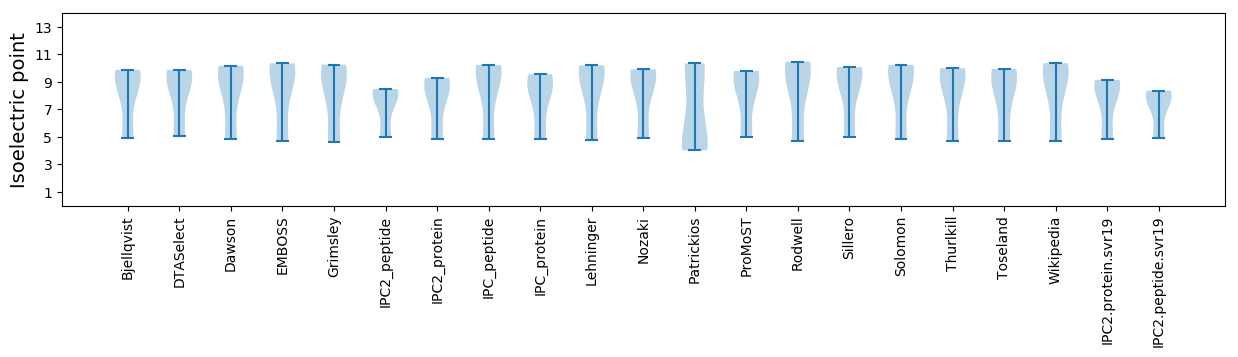
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W9P3|A0A4P8W9P3_9VIRU Uncharacterized protein OS=Tortoise microvirus 39 OX=2583142 PE=4 SV=1
MM1 pKa = 7.58EE2 pKa = 5.55GNMSKK7 pKa = 10.72SNKK10 pKa = 8.22EE11 pKa = 3.32RR12 pKa = 11.84QAQYY16 pKa = 10.27RR17 pKa = 11.84AKK19 pKa = 10.56LEE21 pKa = 4.27SLGQTNVTGIVNYY34 pKa = 8.01TQVGDD39 pKa = 3.7VLILLRR45 pKa = 11.84RR46 pKa = 11.84LRR48 pKa = 11.84DD49 pKa = 3.7NPDD52 pKa = 3.39LEE54 pKa = 4.51VGLIRR59 pKa = 11.84NTRR62 pKa = 11.84TGRR65 pKa = 11.84YY66 pKa = 7.34EE67 pKa = 3.79KK68 pKa = 10.96LL69 pKa = 3.29
MM1 pKa = 7.58EE2 pKa = 5.55GNMSKK7 pKa = 10.72SNKK10 pKa = 8.22EE11 pKa = 3.32RR12 pKa = 11.84QAQYY16 pKa = 10.27RR17 pKa = 11.84AKK19 pKa = 10.56LEE21 pKa = 4.27SLGQTNVTGIVNYY34 pKa = 8.01TQVGDD39 pKa = 3.7VLILLRR45 pKa = 11.84RR46 pKa = 11.84LRR48 pKa = 11.84DD49 pKa = 3.7NPDD52 pKa = 3.39LEE54 pKa = 4.51VGLIRR59 pKa = 11.84NTRR62 pKa = 11.84TGRR65 pKa = 11.84YY66 pKa = 7.34EE67 pKa = 3.79KK68 pKa = 10.96LL69 pKa = 3.29
Molecular weight: 7.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1717 |
67 |
548 |
245.3 |
27.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.25 ± 0.836 | 0.815 ± 0.43 |
5.649 ± 0.364 | 5.591 ± 0.684 |
3.844 ± 0.372 | 7.804 ± 0.775 |
2.854 ± 0.616 | 3.378 ± 0.328 |
3.902 ± 0.365 | 7.921 ± 0.476 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.97 ± 0.289 | 4.601 ± 0.497 |
5.591 ± 0.592 | 4.252 ± 0.349 |
7.338 ± 0.77 | 4.543 ± 0.535 |
6.057 ± 0.5 | 7.455 ± 0.735 |
1.864 ± 0.381 | 3.32 ± 0.792 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
