
Torque teno virus 26
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 7.63
Get precalculated fractions of proteins
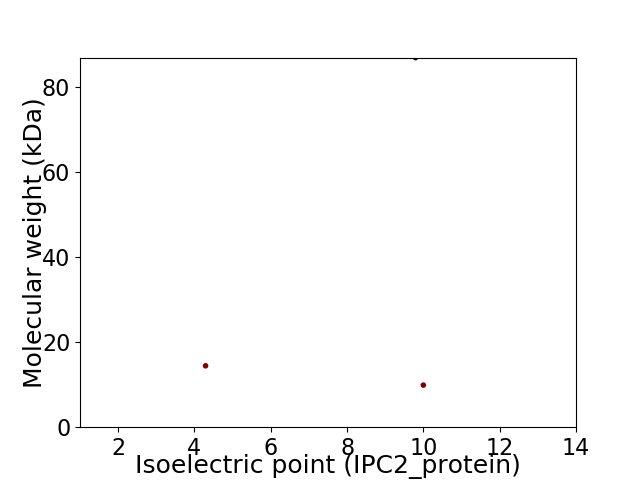
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9DUC8|Q9DUC8_9VIRU Uncharacterized protein OS=Torque teno virus 26 OX=687365 PE=4 SV=2
MM1 pKa = 7.72AEE3 pKa = 4.29APTGQWEE10 pKa = 4.26PPQQGVGPRR19 pKa = 11.84EE20 pKa = 3.86LDD22 pKa = 2.77WWRR25 pKa = 11.84GTWWNHH31 pKa = 5.08AAFCGCGDD39 pKa = 3.88PSFHH43 pKa = 7.13LALLSHH49 pKa = 6.91HH50 pKa = 6.49NFGPRR55 pKa = 11.84GPPGGPPAPPPPPPPVAQRR74 pKa = 11.84GPPNDD79 pKa = 3.77PGLPRR84 pKa = 11.84FLPLPGLPPQPPEE97 pKa = 4.76DD98 pKa = 3.81PPARR102 pKa = 11.84PGGGGDD108 pKa = 3.81GPDD111 pKa = 3.25AAVGGGGGAEE121 pKa = 4.38DD122 pKa = 3.68AWDD125 pKa = 4.28PGDD128 pKa = 4.29VEE130 pKa = 4.55EE131 pKa = 5.18LLAAAAEE138 pKa = 4.41EE139 pKa = 4.41EE140 pKa = 4.73GG141 pKa = 4.05
MM1 pKa = 7.72AEE3 pKa = 4.29APTGQWEE10 pKa = 4.26PPQQGVGPRR19 pKa = 11.84EE20 pKa = 3.86LDD22 pKa = 2.77WWRR25 pKa = 11.84GTWWNHH31 pKa = 5.08AAFCGCGDD39 pKa = 3.88PSFHH43 pKa = 7.13LALLSHH49 pKa = 6.91HH50 pKa = 6.49NFGPRR55 pKa = 11.84GPPGGPPAPPPPPPPVAQRR74 pKa = 11.84GPPNDD79 pKa = 3.77PGLPRR84 pKa = 11.84FLPLPGLPPQPPEE97 pKa = 4.76DD98 pKa = 3.81PPARR102 pKa = 11.84PGGGGDD108 pKa = 3.81GPDD111 pKa = 3.25AAVGGGGGAEE121 pKa = 4.38DD122 pKa = 3.68AWDD125 pKa = 4.28PGDD128 pKa = 4.29VEE130 pKa = 4.55EE131 pKa = 5.18LLAAAAEE138 pKa = 4.41EE139 pKa = 4.41EE140 pKa = 4.73GG141 pKa = 4.05
Molecular weight: 14.43 kDa
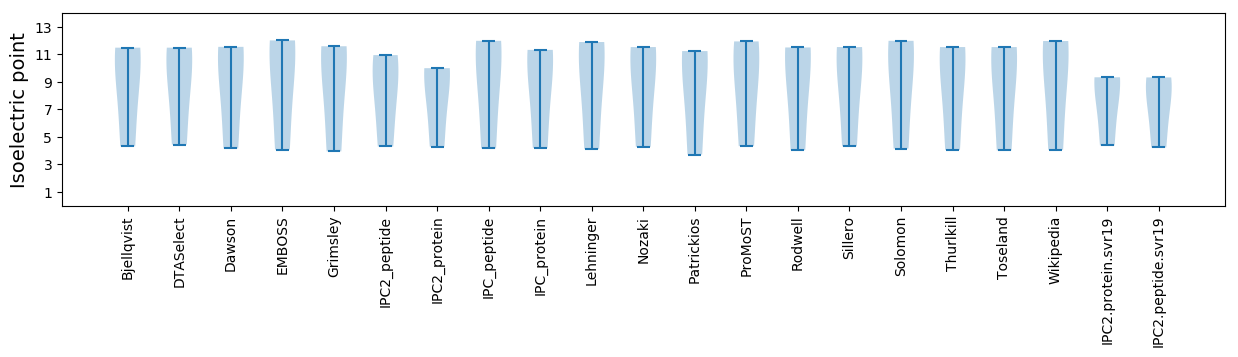
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9DUC8|Q9DUC8_9VIRU Uncharacterized protein OS=Torque teno virus 26 OX=687365 PE=4 SV=2
MM1 pKa = 7.08AWWWRR6 pKa = 11.84RR7 pKa = 11.84WPRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84WRR16 pKa = 11.84RR17 pKa = 11.84WGRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84VGSWRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84GVAGRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84RR40 pKa = 11.84VRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84VWRR51 pKa = 11.84RR52 pKa = 11.84GRR54 pKa = 11.84RR55 pKa = 11.84WRR57 pKa = 11.84LRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84LRR65 pKa = 11.84RR66 pKa = 11.84YY67 pKa = 8.87FRR69 pKa = 11.84NTLILRR75 pKa = 11.84QWQPTTIKK83 pKa = 10.35SLRR86 pKa = 11.84ISGWFPGVVCSNRR99 pKa = 11.84RR100 pKa = 11.84SQNNYY105 pKa = 8.73IVHH108 pKa = 6.68LPDD111 pKa = 3.15IPARR115 pKa = 11.84GGGFGGNISITKK127 pKa = 9.16WNLSMLWHH135 pKa = 6.65EE136 pKa = 4.34NLMNRR141 pKa = 11.84NRR143 pKa = 11.84WSRR146 pKa = 11.84RR147 pKa = 11.84NNDD150 pKa = 3.26LDD152 pKa = 3.71LVRR155 pKa = 11.84YY156 pKa = 9.96LGGSWKK162 pKa = 10.37FYY164 pKa = 10.27RR165 pKa = 11.84DD166 pKa = 3.32PDD168 pKa = 3.51RR169 pKa = 11.84DD170 pKa = 4.35FIATYY175 pKa = 10.46SLEE178 pKa = 4.34SPMTTNQYY186 pKa = 10.58SHH188 pKa = 6.69LQSHH192 pKa = 6.04PQIMLLRR199 pKa = 11.84RR200 pKa = 11.84HH201 pKa = 6.79RR202 pKa = 11.84ILIPSLKK209 pKa = 9.01TKK211 pKa = 10.26PRR213 pKa = 11.84GKK215 pKa = 9.67PYY217 pKa = 10.61VKK219 pKa = 10.25RR220 pKa = 11.84KK221 pKa = 9.12FKK223 pKa = 10.38PPKK226 pKa = 9.85LMRR229 pKa = 11.84NQWYY233 pKa = 8.74FQAHH237 pKa = 5.87FCNVNLIKK245 pKa = 10.38LTVVGLDD252 pKa = 3.66LQKK255 pKa = 10.82AWLRR259 pKa = 11.84RR260 pKa = 11.84GTEE263 pKa = 3.84SPIAEE268 pKa = 4.05FAVLQQSLYY277 pKa = 11.26NNISLTATEE286 pKa = 4.61TNSEE290 pKa = 4.17EE291 pKa = 4.97QKK293 pKa = 9.71KK294 pKa = 6.97TWEE297 pKa = 4.28NVWAFPMSMTMNFRR311 pKa = 11.84EE312 pKa = 4.16LLKK315 pKa = 10.26TLGATDD321 pKa = 4.87AEE323 pKa = 4.66LKK325 pKa = 10.65DD326 pKa = 3.72GPKK329 pKa = 9.65RR330 pKa = 11.84DD331 pKa = 3.88SVPTLWKK338 pKa = 10.48KK339 pKa = 10.64YY340 pKa = 6.23VTSNKK345 pKa = 6.76MTEE348 pKa = 3.98NVFNDD353 pKa = 3.58RR354 pKa = 11.84QKK356 pKa = 11.02KK357 pKa = 9.6INWLVSNSGITTTNTLSRR375 pKa = 11.84TMYY378 pKa = 10.25FDD380 pKa = 5.58RR381 pKa = 11.84ICGMFSSYY389 pKa = 11.42VLDD392 pKa = 4.2DD393 pKa = 3.49TVRR396 pKa = 11.84WDD398 pKa = 3.5GTLKK402 pKa = 10.36KK403 pKa = 10.34AYY405 pKa = 10.3VGVRR409 pKa = 11.84YY410 pKa = 9.98NPLIDD415 pKa = 3.99EE416 pKa = 4.48GTGNKK421 pKa = 7.79VWIDD425 pKa = 3.49PVTKK429 pKa = 10.14KK430 pKa = 9.44DD431 pKa = 3.54TKK433 pKa = 10.33FQPPQSLVLLEE444 pKa = 4.75GQPLWLLLFGYY455 pKa = 8.58TDD457 pKa = 4.2WIKK460 pKa = 11.11KK461 pKa = 9.86FYY463 pKa = 10.74ADD465 pKa = 4.31RR466 pKa = 11.84NPGTTYY472 pKa = 10.74RR473 pKa = 11.84VTLLSPWTYY482 pKa = 10.94PKK484 pKa = 9.76LTNKK488 pKa = 10.1DD489 pKa = 3.06LYY491 pKa = 11.11GYY493 pKa = 10.43VPLGDD498 pKa = 4.22DD499 pKa = 4.5FCAGRR504 pKa = 11.84HH505 pKa = 6.11PYY507 pKa = 9.43RR508 pKa = 11.84QYY510 pKa = 11.3KK511 pKa = 7.73ITPEE515 pKa = 4.23WEE517 pKa = 4.27TLWYY521 pKa = 10.12PMVFNQEE528 pKa = 3.87PALEE532 pKa = 4.96AIVNCGPWMPRR543 pKa = 11.84DD544 pKa = 3.34EE545 pKa = 4.73EE546 pKa = 4.12ARR548 pKa = 11.84SWQLNLGYY556 pKa = 9.4QFRR559 pKa = 11.84FKK561 pKa = 10.88LGGHH565 pKa = 7.3LPPGQPPEE573 pKa = 4.84DD574 pKa = 3.94PCKK577 pKa = 10.6QPTHH581 pKa = 7.07DD582 pKa = 4.32LPEE585 pKa = 4.3TNMLQLAVQASNPRR599 pKa = 11.84TVDD602 pKa = 3.48EE603 pKa = 5.27PFHH606 pKa = 6.83GWDD609 pKa = 3.33LRR611 pKa = 11.84RR612 pKa = 11.84GMFSASSIKK621 pKa = 10.27RR622 pKa = 11.84MRR624 pKa = 11.84EE625 pKa = 3.71YY626 pKa = 10.08QTDD629 pKa = 3.65DD630 pKa = 3.56EE631 pKa = 4.91NFPDD635 pKa = 4.14SPAKK639 pKa = 10.04RR640 pKa = 11.84SRR642 pKa = 11.84YY643 pKa = 9.59DD644 pKa = 3.22PPTEE648 pKa = 4.28GEE650 pKa = 4.13PGALPSGSLQALKK663 pKa = 10.87ALFEE667 pKa = 4.56TPQTPGPLSPSWAKK681 pKa = 10.67ADD683 pKa = 3.71EE684 pKa = 4.47APEE687 pKa = 4.0VLQLQLQRR695 pKa = 11.84EE696 pKa = 4.52LQRR699 pKa = 11.84QRR701 pKa = 11.84KK702 pKa = 5.62QQQRR706 pKa = 11.84LQQGIQEE713 pKa = 4.62MLLSMKK719 pKa = 8.81LTQMGHH725 pKa = 6.49HH726 pKa = 6.86IDD728 pKa = 3.62HH729 pKa = 7.2RR730 pKa = 11.84LLL732 pKa = 5.03
MM1 pKa = 7.08AWWWRR6 pKa = 11.84RR7 pKa = 11.84WPRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84WRR16 pKa = 11.84RR17 pKa = 11.84WGRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84VGSWRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84GVAGRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84RR40 pKa = 11.84VRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84VWRR51 pKa = 11.84RR52 pKa = 11.84GRR54 pKa = 11.84RR55 pKa = 11.84WRR57 pKa = 11.84LRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84LRR65 pKa = 11.84RR66 pKa = 11.84YY67 pKa = 8.87FRR69 pKa = 11.84NTLILRR75 pKa = 11.84QWQPTTIKK83 pKa = 10.35SLRR86 pKa = 11.84ISGWFPGVVCSNRR99 pKa = 11.84RR100 pKa = 11.84SQNNYY105 pKa = 8.73IVHH108 pKa = 6.68LPDD111 pKa = 3.15IPARR115 pKa = 11.84GGGFGGNISITKK127 pKa = 9.16WNLSMLWHH135 pKa = 6.65EE136 pKa = 4.34NLMNRR141 pKa = 11.84NRR143 pKa = 11.84WSRR146 pKa = 11.84RR147 pKa = 11.84NNDD150 pKa = 3.26LDD152 pKa = 3.71LVRR155 pKa = 11.84YY156 pKa = 9.96LGGSWKK162 pKa = 10.37FYY164 pKa = 10.27RR165 pKa = 11.84DD166 pKa = 3.32PDD168 pKa = 3.51RR169 pKa = 11.84DD170 pKa = 4.35FIATYY175 pKa = 10.46SLEE178 pKa = 4.34SPMTTNQYY186 pKa = 10.58SHH188 pKa = 6.69LQSHH192 pKa = 6.04PQIMLLRR199 pKa = 11.84RR200 pKa = 11.84HH201 pKa = 6.79RR202 pKa = 11.84ILIPSLKK209 pKa = 9.01TKK211 pKa = 10.26PRR213 pKa = 11.84GKK215 pKa = 9.67PYY217 pKa = 10.61VKK219 pKa = 10.25RR220 pKa = 11.84KK221 pKa = 9.12FKK223 pKa = 10.38PPKK226 pKa = 9.85LMRR229 pKa = 11.84NQWYY233 pKa = 8.74FQAHH237 pKa = 5.87FCNVNLIKK245 pKa = 10.38LTVVGLDD252 pKa = 3.66LQKK255 pKa = 10.82AWLRR259 pKa = 11.84RR260 pKa = 11.84GTEE263 pKa = 3.84SPIAEE268 pKa = 4.05FAVLQQSLYY277 pKa = 11.26NNISLTATEE286 pKa = 4.61TNSEE290 pKa = 4.17EE291 pKa = 4.97QKK293 pKa = 9.71KK294 pKa = 6.97TWEE297 pKa = 4.28NVWAFPMSMTMNFRR311 pKa = 11.84EE312 pKa = 4.16LLKK315 pKa = 10.26TLGATDD321 pKa = 4.87AEE323 pKa = 4.66LKK325 pKa = 10.65DD326 pKa = 3.72GPKK329 pKa = 9.65RR330 pKa = 11.84DD331 pKa = 3.88SVPTLWKK338 pKa = 10.48KK339 pKa = 10.64YY340 pKa = 6.23VTSNKK345 pKa = 6.76MTEE348 pKa = 3.98NVFNDD353 pKa = 3.58RR354 pKa = 11.84QKK356 pKa = 11.02KK357 pKa = 9.6INWLVSNSGITTTNTLSRR375 pKa = 11.84TMYY378 pKa = 10.25FDD380 pKa = 5.58RR381 pKa = 11.84ICGMFSSYY389 pKa = 11.42VLDD392 pKa = 4.2DD393 pKa = 3.49TVRR396 pKa = 11.84WDD398 pKa = 3.5GTLKK402 pKa = 10.36KK403 pKa = 10.34AYY405 pKa = 10.3VGVRR409 pKa = 11.84YY410 pKa = 9.98NPLIDD415 pKa = 3.99EE416 pKa = 4.48GTGNKK421 pKa = 7.79VWIDD425 pKa = 3.49PVTKK429 pKa = 10.14KK430 pKa = 9.44DD431 pKa = 3.54TKK433 pKa = 10.33FQPPQSLVLLEE444 pKa = 4.75GQPLWLLLFGYY455 pKa = 8.58TDD457 pKa = 4.2WIKK460 pKa = 11.11KK461 pKa = 9.86FYY463 pKa = 10.74ADD465 pKa = 4.31RR466 pKa = 11.84NPGTTYY472 pKa = 10.74RR473 pKa = 11.84VTLLSPWTYY482 pKa = 10.94PKK484 pKa = 9.76LTNKK488 pKa = 10.1DD489 pKa = 3.06LYY491 pKa = 11.11GYY493 pKa = 10.43VPLGDD498 pKa = 4.22DD499 pKa = 4.5FCAGRR504 pKa = 11.84HH505 pKa = 6.11PYY507 pKa = 9.43RR508 pKa = 11.84QYY510 pKa = 11.3KK511 pKa = 7.73ITPEE515 pKa = 4.23WEE517 pKa = 4.27TLWYY521 pKa = 10.12PMVFNQEE528 pKa = 3.87PALEE532 pKa = 4.96AIVNCGPWMPRR543 pKa = 11.84DD544 pKa = 3.34EE545 pKa = 4.73EE546 pKa = 4.12ARR548 pKa = 11.84SWQLNLGYY556 pKa = 9.4QFRR559 pKa = 11.84FKK561 pKa = 10.88LGGHH565 pKa = 7.3LPPGQPPEE573 pKa = 4.84DD574 pKa = 3.94PCKK577 pKa = 10.6QPTHH581 pKa = 7.07DD582 pKa = 4.32LPEE585 pKa = 4.3TNMLQLAVQASNPRR599 pKa = 11.84TVDD602 pKa = 3.48EE603 pKa = 5.27PFHH606 pKa = 6.83GWDD609 pKa = 3.33LRR611 pKa = 11.84RR612 pKa = 11.84GMFSASSIKK621 pKa = 10.27RR622 pKa = 11.84MRR624 pKa = 11.84EE625 pKa = 3.71YY626 pKa = 10.08QTDD629 pKa = 3.65DD630 pKa = 3.56EE631 pKa = 4.91NFPDD635 pKa = 4.14SPAKK639 pKa = 10.04RR640 pKa = 11.84SRR642 pKa = 11.84YY643 pKa = 9.59DD644 pKa = 3.22PPTEE648 pKa = 4.28GEE650 pKa = 4.13PGALPSGSLQALKK663 pKa = 10.87ALFEE667 pKa = 4.56TPQTPGPLSPSWAKK681 pKa = 10.67ADD683 pKa = 3.71EE684 pKa = 4.47APEE687 pKa = 4.0VLQLQLQRR695 pKa = 11.84EE696 pKa = 4.52LQRR699 pKa = 11.84QRR701 pKa = 11.84KK702 pKa = 5.62QQQRR706 pKa = 11.84LQQGIQEE713 pKa = 4.62MLLSMKK719 pKa = 8.81LTQMGHH725 pKa = 6.49HH726 pKa = 6.86IDD728 pKa = 3.62HH729 pKa = 7.2RR730 pKa = 11.84LLL732 pKa = 5.03
Molecular weight: 86.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
960 |
87 |
732 |
320.0 |
37.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.0 ± 1.937 | 1.042 ± 0.419 |
4.792 ± 0.515 | 4.688 ± 0.739 |
3.229 ± 0.331 | 7.5 ± 3.023 |
1.771 ± 0.507 | 2.708 ± 1.082 |
5.104 ± 1.627 | 9.271 ± 1.403 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.396 ± 0.535 | 4.375 ± 0.829 |
9.271 ± 3.947 | 5.104 ± 0.456 |
11.146 ± 2.136 | 6.563 ± 4.789 |
5.417 ± 1.342 | 3.854 ± 0.939 |
3.958 ± 1.118 | 2.813 ± 1.132 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
