
CRESS virus sp. ctBNR11
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; unclassified Cressdnaviricota; CRESS viruses
Average proteome isoelectric point is 8.52
Get precalculated fractions of proteins
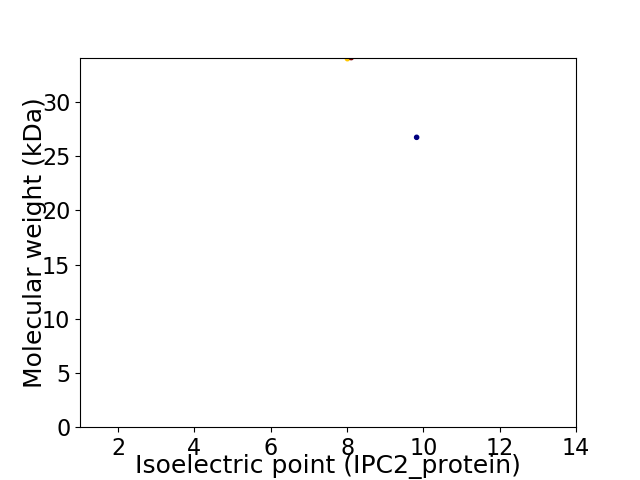
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W8D9|A0A5Q2W8D9_9VIRU Rep protein OS=CRESS virus sp. ctBNR11 OX=2656677 PE=4 SV=1
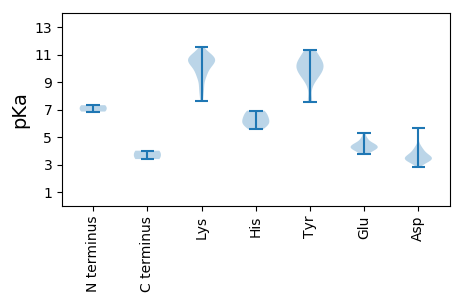
MM1 pKa = 7.32AAEE4 pKa = 4.32TEE6 pKa = 4.54GTNGTVEE13 pKa = 4.48NIGNRR18 pKa = 11.84NKK20 pKa = 10.12VLNQRR25 pKa = 11.84NRR27 pKa = 11.84SRR29 pKa = 11.84AWCFTKK35 pKa = 10.81DD36 pKa = 3.17NATKK40 pKa = 10.42EE41 pKa = 4.11DD42 pKa = 3.44VGTFIKK48 pKa = 10.67YY49 pKa = 9.09FQLEE53 pKa = 4.13DD54 pKa = 3.57MDD56 pKa = 5.68YY57 pKa = 9.55MFQMEE62 pKa = 4.53KK63 pKa = 10.96GEE65 pKa = 4.17KK66 pKa = 9.11TGMIHH71 pKa = 6.5LQGVCRR77 pKa = 11.84FKK79 pKa = 11.02NPRR82 pKa = 11.84DD83 pKa = 3.12RR84 pKa = 11.84WPEE87 pKa = 3.78LKK89 pKa = 10.73AHH91 pKa = 6.88WDD93 pKa = 3.47RR94 pKa = 11.84CRR96 pKa = 11.84SWKK99 pKa = 10.31KK100 pKa = 10.71SIIYY104 pKa = 9.7CSKK107 pKa = 8.49TDD109 pKa = 3.46TRR111 pKa = 11.84IEE113 pKa = 4.34GPWTNLDD120 pKa = 3.3IKK122 pKa = 11.03VKK124 pKa = 10.34EE125 pKa = 4.18KK126 pKa = 10.47PQDD129 pKa = 3.58YY130 pKa = 10.98LKK132 pKa = 11.06DD133 pKa = 3.47KK134 pKa = 10.58EE135 pKa = 4.87LYY137 pKa = 9.55PFQAEE142 pKa = 3.88ILMMLGGKK150 pKa = 9.72IMDD153 pKa = 3.97RR154 pKa = 11.84TIFWYY159 pKa = 10.39FDD161 pKa = 3.32EE162 pKa = 5.27KK163 pKa = 11.56GNTGKK168 pKa = 8.5STLVRR173 pKa = 11.84HH174 pKa = 6.24ILLNNQAICRR184 pKa = 11.84SGSGKK189 pKa = 10.18DD190 pKa = 3.46CYY192 pKa = 10.62CALKK196 pKa = 10.49LRR198 pKa = 11.84CEE200 pKa = 4.44DD201 pKa = 3.32NKK203 pKa = 10.9EE204 pKa = 3.92PNIVIFDD211 pKa = 3.87LTRR214 pKa = 11.84DD215 pKa = 3.71EE216 pKa = 4.71EE217 pKa = 4.47NKK219 pKa = 10.42CNYY222 pKa = 9.7KK223 pKa = 10.65VIEE226 pKa = 4.45TIKK229 pKa = 10.94NGVFFSGKK237 pKa = 9.99YY238 pKa = 7.59EE239 pKa = 4.22SKK241 pKa = 10.72DD242 pKa = 3.23IIINPPHH249 pKa = 6.92VIVFSNWKK257 pKa = 9.44PKK259 pKa = 9.6IDD261 pKa = 4.2KK262 pKa = 10.22LSKK265 pKa = 10.37DD266 pKa = 2.8RR267 pKa = 11.84WSIHH271 pKa = 5.56EE272 pKa = 3.96IKK274 pKa = 10.69RR275 pKa = 11.84PDD277 pKa = 3.47FSSYY281 pKa = 10.46PIYY284 pKa = 10.3IEE286 pKa = 4.24NMM288 pKa = 3.4
MM1 pKa = 7.32AAEE4 pKa = 4.32TEE6 pKa = 4.54GTNGTVEE13 pKa = 4.48NIGNRR18 pKa = 11.84NKK20 pKa = 10.12VLNQRR25 pKa = 11.84NRR27 pKa = 11.84SRR29 pKa = 11.84AWCFTKK35 pKa = 10.81DD36 pKa = 3.17NATKK40 pKa = 10.42EE41 pKa = 4.11DD42 pKa = 3.44VGTFIKK48 pKa = 10.67YY49 pKa = 9.09FQLEE53 pKa = 4.13DD54 pKa = 3.57MDD56 pKa = 5.68YY57 pKa = 9.55MFQMEE62 pKa = 4.53KK63 pKa = 10.96GEE65 pKa = 4.17KK66 pKa = 9.11TGMIHH71 pKa = 6.5LQGVCRR77 pKa = 11.84FKK79 pKa = 11.02NPRR82 pKa = 11.84DD83 pKa = 3.12RR84 pKa = 11.84WPEE87 pKa = 3.78LKK89 pKa = 10.73AHH91 pKa = 6.88WDD93 pKa = 3.47RR94 pKa = 11.84CRR96 pKa = 11.84SWKK99 pKa = 10.31KK100 pKa = 10.71SIIYY104 pKa = 9.7CSKK107 pKa = 8.49TDD109 pKa = 3.46TRR111 pKa = 11.84IEE113 pKa = 4.34GPWTNLDD120 pKa = 3.3IKK122 pKa = 11.03VKK124 pKa = 10.34EE125 pKa = 4.18KK126 pKa = 10.47PQDD129 pKa = 3.58YY130 pKa = 10.98LKK132 pKa = 11.06DD133 pKa = 3.47KK134 pKa = 10.58EE135 pKa = 4.87LYY137 pKa = 9.55PFQAEE142 pKa = 3.88ILMMLGGKK150 pKa = 9.72IMDD153 pKa = 3.97RR154 pKa = 11.84TIFWYY159 pKa = 10.39FDD161 pKa = 3.32EE162 pKa = 5.27KK163 pKa = 11.56GNTGKK168 pKa = 8.5STLVRR173 pKa = 11.84HH174 pKa = 6.24ILLNNQAICRR184 pKa = 11.84SGSGKK189 pKa = 10.18DD190 pKa = 3.46CYY192 pKa = 10.62CALKK196 pKa = 10.49LRR198 pKa = 11.84CEE200 pKa = 4.44DD201 pKa = 3.32NKK203 pKa = 10.9EE204 pKa = 3.92PNIVIFDD211 pKa = 3.87LTRR214 pKa = 11.84DD215 pKa = 3.71EE216 pKa = 4.71EE217 pKa = 4.47NKK219 pKa = 10.42CNYY222 pKa = 9.7KK223 pKa = 10.65VIEE226 pKa = 4.45TIKK229 pKa = 10.94NGVFFSGKK237 pKa = 9.99YY238 pKa = 7.59EE239 pKa = 4.22SKK241 pKa = 10.72DD242 pKa = 3.23IIINPPHH249 pKa = 6.92VIVFSNWKK257 pKa = 9.44PKK259 pKa = 9.6IDD261 pKa = 4.2KK262 pKa = 10.22LSKK265 pKa = 10.37DD266 pKa = 2.8RR267 pKa = 11.84WSIHH271 pKa = 5.56EE272 pKa = 3.96IKK274 pKa = 10.69RR275 pKa = 11.84PDD277 pKa = 3.47FSSYY281 pKa = 10.46PIYY284 pKa = 10.3IEE286 pKa = 4.24NMM288 pKa = 3.4
Molecular weight: 33.97 kDa
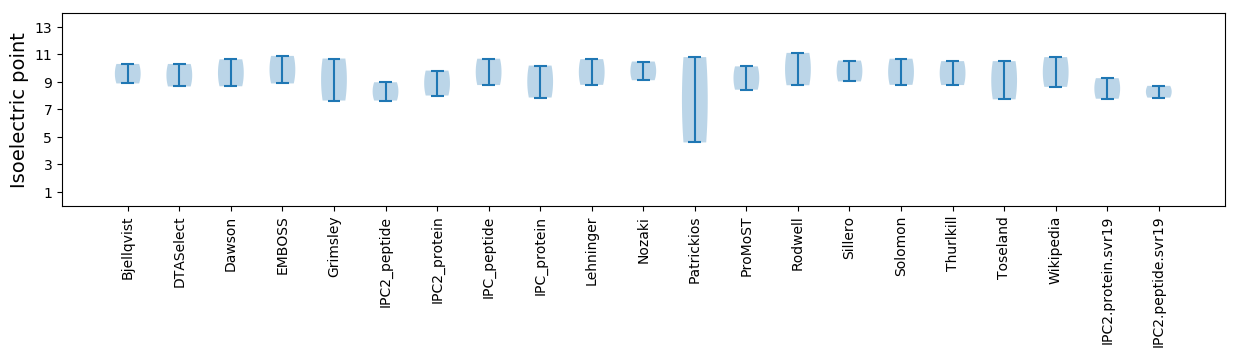
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W8D9|A0A5Q2W8D9_9VIRU Rep protein OS=CRESS virus sp. ctBNR11 OX=2656677 PE=4 SV=1
MM1 pKa = 6.85YY2 pKa = 10.08RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 8.98SRR8 pKa = 11.84ARR10 pKa = 11.84RR11 pKa = 11.84VRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FMNKK19 pKa = 7.64WGAKK23 pKa = 8.7SGRR26 pKa = 11.84TGWQKK31 pKa = 10.59FKK33 pKa = 10.96YY34 pKa = 10.07AAGKK38 pKa = 7.61TLNFASKK45 pKa = 10.08ALKK48 pKa = 9.6VAHH51 pKa = 5.81SVKK54 pKa = 10.73NLINVEE60 pKa = 4.17WKK62 pKa = 10.66RR63 pKa = 11.84KK64 pKa = 8.08DD65 pKa = 3.33TSSLGGAVGNVSPSTIGGMCVIAQGTDD92 pKa = 3.13YY93 pKa = 11.06NQRR96 pKa = 11.84NGNQVLLKK104 pKa = 10.58QFGLKK109 pKa = 8.45GTLLWNSNSTTQRR122 pKa = 11.84VRR124 pKa = 11.84IVIVMQKK131 pKa = 9.92NAEE134 pKa = 4.23DD135 pKa = 4.01PSTYY139 pKa = 9.61IDD141 pKa = 3.65MYY143 pKa = 11.32GAGLDD148 pKa = 3.41GFRR151 pKa = 11.84YY152 pKa = 10.31VEE154 pKa = 4.92DD155 pKa = 3.82KK156 pKa = 10.31PKK158 pKa = 10.91SRR160 pKa = 11.84IIFSRR165 pKa = 11.84LYY167 pKa = 10.44ILDD170 pKa = 4.05VNHH173 pKa = 6.51KK174 pKa = 9.61QYY176 pKa = 11.31NITFFKK182 pKa = 10.63KK183 pKa = 10.46FYY185 pKa = 7.53KK186 pKa = 9.37THH188 pKa = 6.01MKK190 pKa = 9.43WDD192 pKa = 4.45PNHH195 pKa = 6.01STTSPKK201 pKa = 10.79YY202 pKa = 9.98NDD204 pKa = 3.44FSVLARR210 pKa = 11.84SNDD213 pKa = 3.65DD214 pKa = 3.55DD215 pKa = 4.19NKK217 pKa = 9.42PTFVFNTRR225 pKa = 11.84VTYY228 pKa = 10.3IDD230 pKa = 3.32NN231 pKa = 4.01
MM1 pKa = 6.85YY2 pKa = 10.08RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 8.98SRR8 pKa = 11.84ARR10 pKa = 11.84RR11 pKa = 11.84VRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FMNKK19 pKa = 7.64WGAKK23 pKa = 8.7SGRR26 pKa = 11.84TGWQKK31 pKa = 10.59FKK33 pKa = 10.96YY34 pKa = 10.07AAGKK38 pKa = 7.61TLNFASKK45 pKa = 10.08ALKK48 pKa = 9.6VAHH51 pKa = 5.81SVKK54 pKa = 10.73NLINVEE60 pKa = 4.17WKK62 pKa = 10.66RR63 pKa = 11.84KK64 pKa = 8.08DD65 pKa = 3.33TSSLGGAVGNVSPSTIGGMCVIAQGTDD92 pKa = 3.13YY93 pKa = 11.06NQRR96 pKa = 11.84NGNQVLLKK104 pKa = 10.58QFGLKK109 pKa = 8.45GTLLWNSNSTTQRR122 pKa = 11.84VRR124 pKa = 11.84IVIVMQKK131 pKa = 9.92NAEE134 pKa = 4.23DD135 pKa = 4.01PSTYY139 pKa = 9.61IDD141 pKa = 3.65MYY143 pKa = 11.32GAGLDD148 pKa = 3.41GFRR151 pKa = 11.84YY152 pKa = 10.31VEE154 pKa = 4.92DD155 pKa = 3.82KK156 pKa = 10.31PKK158 pKa = 10.91SRR160 pKa = 11.84IIFSRR165 pKa = 11.84LYY167 pKa = 10.44ILDD170 pKa = 4.05VNHH173 pKa = 6.51KK174 pKa = 9.61QYY176 pKa = 11.31NITFFKK182 pKa = 10.63KK183 pKa = 10.46FYY185 pKa = 7.53KK186 pKa = 9.37THH188 pKa = 6.01MKK190 pKa = 9.43WDD192 pKa = 4.45PNHH195 pKa = 6.01STTSPKK201 pKa = 10.79YY202 pKa = 9.98NDD204 pKa = 3.44FSVLARR210 pKa = 11.84SNDD213 pKa = 3.65DD214 pKa = 3.55DD215 pKa = 4.19NKK217 pKa = 9.42PTFVFNTRR225 pKa = 11.84VTYY228 pKa = 10.3IDD230 pKa = 3.32NN231 pKa = 4.01
Molecular weight: 26.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
519 |
231 |
288 |
259.5 |
30.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.854 ± 0.876 | 1.927 ± 0.976 |
6.358 ± 0.477 | 4.624 ± 2.172 |
4.817 ± 0.247 | 6.166 ± 0.497 |
1.734 ± 0.002 | 6.936 ± 1.421 |
10.405 ± 0.575 | 5.588 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.89 ± 0.191 | 7.514 ± 0.464 |
3.276 ± 0.443 | 2.89 ± 0.374 |
6.936 ± 0.842 | 5.973 ± 0.906 |
5.973 ± 0.623 | 5.202 ± 1.126 |
2.505 ± 0.222 | 4.432 ± 0.499 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
