
Xiphophorus maculatus (Southern platyfish) (Platypoecilus maculatus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei;
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
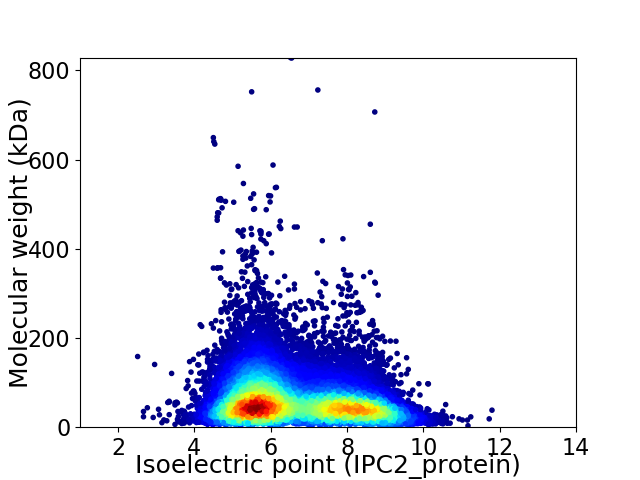
Virtual 2D-PAGE plot for 35279 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
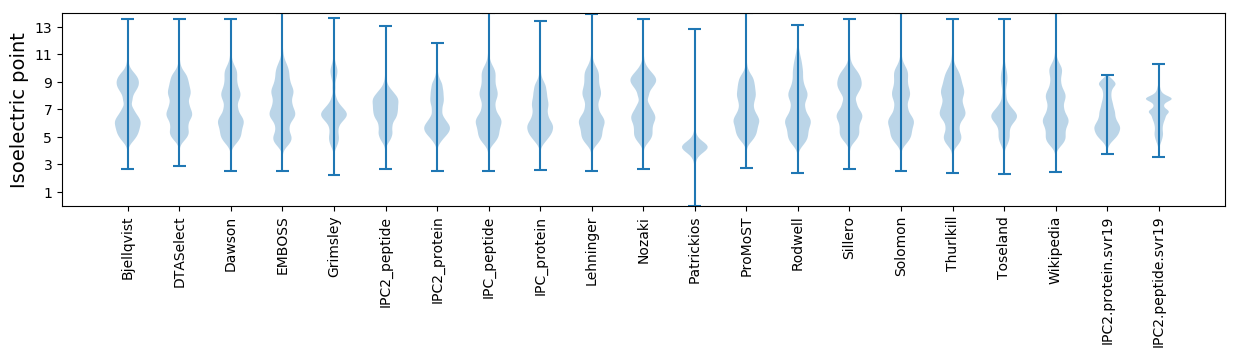
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3B5R7X3|A0A3B5R7X3_XIPMA AdoHcyase_NAD domain-containing protein OS=Xiphophorus maculatus OX=8083 PE=3 SV=1
MM1 pKa = 7.39YY2 pKa = 10.41LEE4 pKa = 4.79NSEE7 pKa = 4.01QGKK10 pKa = 10.25VGGEE14 pKa = 4.4GEE16 pKa = 4.24LSAGAILSGQCAVSKK31 pKa = 8.99VTGVAGLSSSSKK43 pKa = 10.59GSEE46 pKa = 3.88LDD48 pKa = 2.96IKK50 pKa = 11.21ALVGYY55 pKa = 7.3TEE57 pKa = 4.21QSVSGGMDD65 pKa = 3.68PPPLCNMNAPQTQLCNDD82 pKa = 3.86CEE84 pKa = 4.31GLTEE88 pKa = 4.16SFEE91 pKa = 4.71EE92 pKa = 4.36YY93 pKa = 10.4SSSNGLFEE101 pKa = 5.59PDD103 pKa = 3.18GALEE107 pKa = 4.12DD108 pKa = 4.49TEE110 pKa = 5.05CSGLSRR116 pKa = 11.84LYY118 pKa = 10.71HH119 pKa = 6.91EE120 pKa = 5.11CTQHH124 pKa = 6.63CEE126 pKa = 3.9CFKK129 pKa = 10.38ACKK132 pKa = 10.03SSEE135 pKa = 3.77HH136 pKa = 6.51CANHH140 pKa = 5.87SLASKK145 pKa = 10.76SSLCCEE151 pKa = 4.39HH152 pKa = 6.8CPKK155 pKa = 10.59HH156 pKa = 6.58LLLFQQCKK164 pKa = 9.97PSDD167 pKa = 3.8QQSDD171 pKa = 3.72SSDD174 pKa = 3.72YY175 pKa = 11.23EE176 pKa = 4.0PDD178 pKa = 3.42EE179 pKa = 5.43LEE181 pKa = 4.29VNSDD185 pKa = 3.4SFGEE189 pKa = 4.09FEE191 pKa = 4.25MPGFVSEE198 pKa = 4.29CTEE201 pKa = 3.89NLMLPLDD208 pKa = 4.15RR209 pKa = 11.84QCEE212 pKa = 4.23DD213 pKa = 3.42SDD215 pKa = 4.47SEE217 pKa = 4.17QDD219 pKa = 3.57RR220 pKa = 11.84STEE223 pKa = 3.75DD224 pKa = 3.09SEE226 pKa = 5.67QNFTQNISDD235 pKa = 3.82SMDD238 pKa = 3.55SLTCAANFSEE248 pKa = 4.41YY249 pKa = 10.82DD250 pKa = 3.46EE251 pKa = 5.39SQDD254 pKa = 3.53NYY256 pKa = 10.81CDD258 pKa = 3.94EE259 pKa = 4.53EE260 pKa = 4.58EE261 pKa = 4.09EE262 pKa = 4.9SYY264 pKa = 11.63DD265 pKa = 3.56HH266 pKa = 7.47NEE268 pKa = 3.96MVSGEE273 pKa = 4.1EE274 pKa = 4.06SSTLSEE280 pKa = 4.55DD281 pKa = 3.98TPGDD285 pKa = 3.51PCEE288 pKa = 4.49SDD290 pKa = 3.16GGEE293 pKa = 4.04EE294 pKa = 4.4NIEE297 pKa = 3.95QTSTGDD303 pKa = 3.03ASEE306 pKa = 5.3DD307 pKa = 3.21VDD309 pKa = 4.06EE310 pKa = 4.33VHH312 pKa = 7.45RR313 pKa = 11.84DD314 pKa = 3.5DD315 pKa = 6.41SEE317 pKa = 5.57DD318 pKa = 3.52EE319 pKa = 4.3SSQHH323 pKa = 6.73GNFEE327 pKa = 4.32SCAEE331 pKa = 4.18EE332 pKa = 4.99DD333 pKa = 4.77LSSDD337 pKa = 3.43DD338 pKa = 3.71SKK340 pKa = 11.92SFKK343 pKa = 10.11TCLDD347 pKa = 3.1NSFPSDD353 pKa = 3.56PCSDD357 pKa = 3.43SSEE360 pKa = 4.42EE361 pKa = 3.96SDD363 pKa = 4.94KK364 pKa = 11.61AALDD368 pKa = 3.86DD369 pKa = 4.32TSDD372 pKa = 4.98EE373 pKa = 4.14PTQWEE378 pKa = 4.39SFEE381 pKa = 4.82EE382 pKa = 3.99NDD384 pKa = 4.61AEE386 pKa = 4.22KK387 pKa = 10.7QPSVEE392 pKa = 4.06KK393 pKa = 10.89SSEE396 pKa = 4.01EE397 pKa = 3.76EE398 pKa = 4.1DD399 pKa = 3.44KK400 pKa = 11.45KK401 pKa = 10.56KK402 pKa = 9.45TSAVNTVIEE411 pKa = 4.88DD412 pKa = 3.71YY413 pKa = 11.25FDD415 pKa = 4.29LFDD418 pKa = 4.52RR419 pKa = 11.84AGCSGQAFAQKK430 pKa = 10.23RR431 pKa = 11.84NYY433 pKa = 9.5ISCFDD438 pKa = 3.83GGDD441 pKa = 3.75VHH443 pKa = 7.22HH444 pKa = 7.04HH445 pKa = 5.92LHH447 pKa = 6.9LEE449 pKa = 4.01QEE451 pKa = 4.23ALKK454 pKa = 10.8QSVTTAHH461 pKa = 6.79KK462 pKa = 10.56EE463 pKa = 3.75EE464 pKa = 4.15TKK466 pKa = 10.75EE467 pKa = 4.11VIEE470 pKa = 4.11EE471 pKa = 4.09QEE473 pKa = 4.14KK474 pKa = 10.72DD475 pKa = 3.52VHH477 pKa = 5.87TGEE480 pKa = 4.25VTEE483 pKa = 4.67TNSQEE488 pKa = 4.1KK489 pKa = 10.57DD490 pKa = 3.32EE491 pKa = 4.67HH492 pKa = 8.14CEE494 pKa = 3.83DD495 pKa = 5.18SEE497 pKa = 5.44EE498 pKa = 4.12EE499 pKa = 4.35DD500 pKa = 3.99LNQRR504 pKa = 11.84DD505 pKa = 3.6TGSYY509 pKa = 10.8DD510 pKa = 3.42SEE512 pKa = 3.9NWTLKK517 pKa = 10.65SEE519 pKa = 4.2SSSTKK524 pKa = 10.73GEE526 pKa = 4.12AEE528 pKa = 4.0DD529 pKa = 4.07NEE531 pKa = 4.5PFGLYY536 pKa = 9.63PEE538 pKa = 5.13YY539 pKa = 11.01YY540 pKa = 9.54QDD542 pKa = 5.49SEE544 pKa = 5.29DD545 pKa = 3.25EE546 pKa = 4.15MYY548 pKa = 11.45YY549 pKa = 10.83NDD551 pKa = 4.41EE552 pKa = 3.65AWGYY556 pKa = 10.67DD557 pKa = 3.46GQISDD562 pKa = 5.53DD563 pKa = 3.72YY564 pKa = 11.8DD565 pKa = 3.86EE566 pKa = 5.45EE567 pKa = 5.21PEE569 pKa = 4.39SYY571 pKa = 10.72LDD573 pKa = 3.57SFRR576 pKa = 11.84AEE578 pKa = 4.07STFAPCAQEE587 pKa = 4.11IFVEE591 pKa = 4.55GDD593 pKa = 3.12VCEE596 pKa = 5.88DD597 pKa = 3.24IDD599 pKa = 5.57SVGEE603 pKa = 4.52DD604 pKa = 2.91IDD606 pKa = 4.63LVGEE610 pKa = 4.46VKK612 pKa = 10.67SKK614 pKa = 11.16ALTEE618 pKa = 4.16TTLDD622 pKa = 3.78LKK624 pKa = 11.23DD625 pKa = 4.18NKK627 pKa = 10.75NSDD630 pKa = 3.57AEE632 pKa = 4.22LDD634 pKa = 3.76GFSAFSEE641 pKa = 4.27MEE643 pKa = 4.41PYY645 pKa = 9.67WGLTEE650 pKa = 4.02EE651 pKa = 4.36QTRR654 pKa = 11.84EE655 pKa = 4.1CFPSGVEE662 pKa = 4.77DD663 pKa = 4.16YY664 pKa = 10.65YY665 pKa = 11.5AYY667 pKa = 10.35QIRR670 pKa = 11.84SIQSSDD676 pKa = 3.04ARR678 pKa = 11.84PLSRR682 pKa = 11.84FILDD686 pKa = 3.49NRR688 pKa = 11.84SSDD691 pKa = 3.33GKK693 pKa = 10.84AVGDD697 pKa = 3.71QGPEE701 pKa = 3.89DD702 pKa = 4.0VEE704 pKa = 4.24EE705 pKa = 4.47SSALLGINVLEE716 pKa = 5.28LIRR719 pKa = 11.84KK720 pKa = 6.27EE721 pKa = 3.95QSLEE725 pKa = 4.08PEE727 pKa = 4.35NEE729 pKa = 3.89LQGISDD735 pKa = 4.72LSPPLGFIHH744 pKa = 6.78SVASQHH750 pKa = 5.95EE751 pKa = 4.41ATPSQDD757 pKa = 3.55SEE759 pKa = 4.57EE760 pKa = 4.2EE761 pKa = 3.96QSDD764 pKa = 3.95DD765 pKa = 3.93EE766 pKa = 4.63SVEE769 pKa = 3.99NCEE772 pKa = 4.88CEE774 pKa = 4.22YY775 pKa = 10.7CIPPTQQVPVKK786 pKa = 9.76PLLSQSASDD795 pKa = 5.01DD796 pKa = 3.62PAKK799 pKa = 10.35ICVVIDD805 pKa = 3.67LDD807 pKa = 3.74EE808 pKa = 4.47TLVHH812 pKa = 7.13SSFKK816 pKa = 10.34PLNNADD822 pKa = 4.24FIIPVEE828 pKa = 3.94IEE830 pKa = 4.53GIVHH834 pKa = 6.02QVGTT838 pKa = 4.16
MM1 pKa = 7.39YY2 pKa = 10.41LEE4 pKa = 4.79NSEE7 pKa = 4.01QGKK10 pKa = 10.25VGGEE14 pKa = 4.4GEE16 pKa = 4.24LSAGAILSGQCAVSKK31 pKa = 8.99VTGVAGLSSSSKK43 pKa = 10.59GSEE46 pKa = 3.88LDD48 pKa = 2.96IKK50 pKa = 11.21ALVGYY55 pKa = 7.3TEE57 pKa = 4.21QSVSGGMDD65 pKa = 3.68PPPLCNMNAPQTQLCNDD82 pKa = 3.86CEE84 pKa = 4.31GLTEE88 pKa = 4.16SFEE91 pKa = 4.71EE92 pKa = 4.36YY93 pKa = 10.4SSSNGLFEE101 pKa = 5.59PDD103 pKa = 3.18GALEE107 pKa = 4.12DD108 pKa = 4.49TEE110 pKa = 5.05CSGLSRR116 pKa = 11.84LYY118 pKa = 10.71HH119 pKa = 6.91EE120 pKa = 5.11CTQHH124 pKa = 6.63CEE126 pKa = 3.9CFKK129 pKa = 10.38ACKK132 pKa = 10.03SSEE135 pKa = 3.77HH136 pKa = 6.51CANHH140 pKa = 5.87SLASKK145 pKa = 10.76SSLCCEE151 pKa = 4.39HH152 pKa = 6.8CPKK155 pKa = 10.59HH156 pKa = 6.58LLLFQQCKK164 pKa = 9.97PSDD167 pKa = 3.8QQSDD171 pKa = 3.72SSDD174 pKa = 3.72YY175 pKa = 11.23EE176 pKa = 4.0PDD178 pKa = 3.42EE179 pKa = 5.43LEE181 pKa = 4.29VNSDD185 pKa = 3.4SFGEE189 pKa = 4.09FEE191 pKa = 4.25MPGFVSEE198 pKa = 4.29CTEE201 pKa = 3.89NLMLPLDD208 pKa = 4.15RR209 pKa = 11.84QCEE212 pKa = 4.23DD213 pKa = 3.42SDD215 pKa = 4.47SEE217 pKa = 4.17QDD219 pKa = 3.57RR220 pKa = 11.84STEE223 pKa = 3.75DD224 pKa = 3.09SEE226 pKa = 5.67QNFTQNISDD235 pKa = 3.82SMDD238 pKa = 3.55SLTCAANFSEE248 pKa = 4.41YY249 pKa = 10.82DD250 pKa = 3.46EE251 pKa = 5.39SQDD254 pKa = 3.53NYY256 pKa = 10.81CDD258 pKa = 3.94EE259 pKa = 4.53EE260 pKa = 4.58EE261 pKa = 4.09EE262 pKa = 4.9SYY264 pKa = 11.63DD265 pKa = 3.56HH266 pKa = 7.47NEE268 pKa = 3.96MVSGEE273 pKa = 4.1EE274 pKa = 4.06SSTLSEE280 pKa = 4.55DD281 pKa = 3.98TPGDD285 pKa = 3.51PCEE288 pKa = 4.49SDD290 pKa = 3.16GGEE293 pKa = 4.04EE294 pKa = 4.4NIEE297 pKa = 3.95QTSTGDD303 pKa = 3.03ASEE306 pKa = 5.3DD307 pKa = 3.21VDD309 pKa = 4.06EE310 pKa = 4.33VHH312 pKa = 7.45RR313 pKa = 11.84DD314 pKa = 3.5DD315 pKa = 6.41SEE317 pKa = 5.57DD318 pKa = 3.52EE319 pKa = 4.3SSQHH323 pKa = 6.73GNFEE327 pKa = 4.32SCAEE331 pKa = 4.18EE332 pKa = 4.99DD333 pKa = 4.77LSSDD337 pKa = 3.43DD338 pKa = 3.71SKK340 pKa = 11.92SFKK343 pKa = 10.11TCLDD347 pKa = 3.1NSFPSDD353 pKa = 3.56PCSDD357 pKa = 3.43SSEE360 pKa = 4.42EE361 pKa = 3.96SDD363 pKa = 4.94KK364 pKa = 11.61AALDD368 pKa = 3.86DD369 pKa = 4.32TSDD372 pKa = 4.98EE373 pKa = 4.14PTQWEE378 pKa = 4.39SFEE381 pKa = 4.82EE382 pKa = 3.99NDD384 pKa = 4.61AEE386 pKa = 4.22KK387 pKa = 10.7QPSVEE392 pKa = 4.06KK393 pKa = 10.89SSEE396 pKa = 4.01EE397 pKa = 3.76EE398 pKa = 4.1DD399 pKa = 3.44KK400 pKa = 11.45KK401 pKa = 10.56KK402 pKa = 9.45TSAVNTVIEE411 pKa = 4.88DD412 pKa = 3.71YY413 pKa = 11.25FDD415 pKa = 4.29LFDD418 pKa = 4.52RR419 pKa = 11.84AGCSGQAFAQKK430 pKa = 10.23RR431 pKa = 11.84NYY433 pKa = 9.5ISCFDD438 pKa = 3.83GGDD441 pKa = 3.75VHH443 pKa = 7.22HH444 pKa = 7.04HH445 pKa = 5.92LHH447 pKa = 6.9LEE449 pKa = 4.01QEE451 pKa = 4.23ALKK454 pKa = 10.8QSVTTAHH461 pKa = 6.79KK462 pKa = 10.56EE463 pKa = 3.75EE464 pKa = 4.15TKK466 pKa = 10.75EE467 pKa = 4.11VIEE470 pKa = 4.11EE471 pKa = 4.09QEE473 pKa = 4.14KK474 pKa = 10.72DD475 pKa = 3.52VHH477 pKa = 5.87TGEE480 pKa = 4.25VTEE483 pKa = 4.67TNSQEE488 pKa = 4.1KK489 pKa = 10.57DD490 pKa = 3.32EE491 pKa = 4.67HH492 pKa = 8.14CEE494 pKa = 3.83DD495 pKa = 5.18SEE497 pKa = 5.44EE498 pKa = 4.12EE499 pKa = 4.35DD500 pKa = 3.99LNQRR504 pKa = 11.84DD505 pKa = 3.6TGSYY509 pKa = 10.8DD510 pKa = 3.42SEE512 pKa = 3.9NWTLKK517 pKa = 10.65SEE519 pKa = 4.2SSSTKK524 pKa = 10.73GEE526 pKa = 4.12AEE528 pKa = 4.0DD529 pKa = 4.07NEE531 pKa = 4.5PFGLYY536 pKa = 9.63PEE538 pKa = 5.13YY539 pKa = 11.01YY540 pKa = 9.54QDD542 pKa = 5.49SEE544 pKa = 5.29DD545 pKa = 3.25EE546 pKa = 4.15MYY548 pKa = 11.45YY549 pKa = 10.83NDD551 pKa = 4.41EE552 pKa = 3.65AWGYY556 pKa = 10.67DD557 pKa = 3.46GQISDD562 pKa = 5.53DD563 pKa = 3.72YY564 pKa = 11.8DD565 pKa = 3.86EE566 pKa = 5.45EE567 pKa = 5.21PEE569 pKa = 4.39SYY571 pKa = 10.72LDD573 pKa = 3.57SFRR576 pKa = 11.84AEE578 pKa = 4.07STFAPCAQEE587 pKa = 4.11IFVEE591 pKa = 4.55GDD593 pKa = 3.12VCEE596 pKa = 5.88DD597 pKa = 3.24IDD599 pKa = 5.57SVGEE603 pKa = 4.52DD604 pKa = 2.91IDD606 pKa = 4.63LVGEE610 pKa = 4.46VKK612 pKa = 10.67SKK614 pKa = 11.16ALTEE618 pKa = 4.16TTLDD622 pKa = 3.78LKK624 pKa = 11.23DD625 pKa = 4.18NKK627 pKa = 10.75NSDD630 pKa = 3.57AEE632 pKa = 4.22LDD634 pKa = 3.76GFSAFSEE641 pKa = 4.27MEE643 pKa = 4.41PYY645 pKa = 9.67WGLTEE650 pKa = 4.02EE651 pKa = 4.36QTRR654 pKa = 11.84EE655 pKa = 4.1CFPSGVEE662 pKa = 4.77DD663 pKa = 4.16YY664 pKa = 10.65YY665 pKa = 11.5AYY667 pKa = 10.35QIRR670 pKa = 11.84SIQSSDD676 pKa = 3.04ARR678 pKa = 11.84PLSRR682 pKa = 11.84FILDD686 pKa = 3.49NRR688 pKa = 11.84SSDD691 pKa = 3.33GKK693 pKa = 10.84AVGDD697 pKa = 3.71QGPEE701 pKa = 3.89DD702 pKa = 4.0VEE704 pKa = 4.24EE705 pKa = 4.47SSALLGINVLEE716 pKa = 5.28LIRR719 pKa = 11.84KK720 pKa = 6.27EE721 pKa = 3.95QSLEE725 pKa = 4.08PEE727 pKa = 4.35NEE729 pKa = 3.89LQGISDD735 pKa = 4.72LSPPLGFIHH744 pKa = 6.78SVASQHH750 pKa = 5.95EE751 pKa = 4.41ATPSQDD757 pKa = 3.55SEE759 pKa = 4.57EE760 pKa = 4.2EE761 pKa = 3.96QSDD764 pKa = 3.95DD765 pKa = 3.93EE766 pKa = 4.63SVEE769 pKa = 3.99NCEE772 pKa = 4.88CEE774 pKa = 4.22YY775 pKa = 10.7CIPPTQQVPVKK786 pKa = 9.76PLLSQSASDD795 pKa = 5.01DD796 pKa = 3.62PAKK799 pKa = 10.35ICVVIDD805 pKa = 3.67LDD807 pKa = 3.74EE808 pKa = 4.47TLVHH812 pKa = 7.13SSFKK816 pKa = 10.34PLNNADD822 pKa = 4.24FIIPVEE828 pKa = 3.94IEE830 pKa = 4.53GIVHH834 pKa = 6.02QVGTT838 pKa = 4.16
Molecular weight: 92.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3B5QPY3|A0A3B5QPY3_XIPMA Uncharacterized protein OS=Xiphophorus maculatus OX=8083 GN=TENM3 PE=3 SV=1
MM1 pKa = 7.22VVPGLPRR8 pKa = 11.84RR9 pKa = 11.84LQVVPGLPRR18 pKa = 11.84HH19 pKa = 5.5LQVVPGLPRR28 pKa = 11.84RR29 pKa = 11.84FQVVPGLPRR38 pKa = 11.84RR39 pKa = 11.84LQVVPGLPRR48 pKa = 11.84RR49 pKa = 11.84FQVVPGLPRR58 pKa = 11.84RR59 pKa = 11.84LQVVPGLPRR68 pKa = 11.84RR69 pKa = 11.84LQVVPGLPRR78 pKa = 11.84RR79 pKa = 11.84LQVVPGLPRR88 pKa = 11.84RR89 pKa = 11.84LQVVPGLPRR98 pKa = 11.84RR99 pKa = 11.84LQVVPGLPRR108 pKa = 11.84RR109 pKa = 11.84LQVVPGLPRR118 pKa = 11.84RR119 pKa = 11.84FQVVPGLPRR128 pKa = 11.84RR129 pKa = 11.84LQVVPGLPRR138 pKa = 11.84RR139 pKa = 11.84LQVVPGLPRR148 pKa = 11.84RR149 pKa = 11.84LQVVPGLPRR158 pKa = 11.84RR159 pKa = 11.84LQVVPGLPPLEE170 pKa = 4.06MLLRR174 pKa = 11.84AVLHH178 pKa = 6.39RR179 pKa = 11.84ALAVAIHH186 pKa = 6.81LLHH189 pKa = 6.33QVFLGSLSCRR199 pKa = 11.84SVGKK203 pKa = 9.79LAQKK207 pKa = 10.43LL208 pKa = 3.71
MM1 pKa = 7.22VVPGLPRR8 pKa = 11.84RR9 pKa = 11.84LQVVPGLPRR18 pKa = 11.84HH19 pKa = 5.5LQVVPGLPRR28 pKa = 11.84RR29 pKa = 11.84FQVVPGLPRR38 pKa = 11.84RR39 pKa = 11.84LQVVPGLPRR48 pKa = 11.84RR49 pKa = 11.84FQVVPGLPRR58 pKa = 11.84RR59 pKa = 11.84LQVVPGLPRR68 pKa = 11.84RR69 pKa = 11.84LQVVPGLPRR78 pKa = 11.84RR79 pKa = 11.84LQVVPGLPRR88 pKa = 11.84RR89 pKa = 11.84LQVVPGLPRR98 pKa = 11.84RR99 pKa = 11.84LQVVPGLPRR108 pKa = 11.84RR109 pKa = 11.84LQVVPGLPRR118 pKa = 11.84RR119 pKa = 11.84FQVVPGLPRR128 pKa = 11.84RR129 pKa = 11.84LQVVPGLPRR138 pKa = 11.84RR139 pKa = 11.84LQVVPGLPRR148 pKa = 11.84RR149 pKa = 11.84LQVVPGLPRR158 pKa = 11.84RR159 pKa = 11.84LQVVPGLPPLEE170 pKa = 4.06MLLRR174 pKa = 11.84AVLHH178 pKa = 6.39RR179 pKa = 11.84ALAVAIHH186 pKa = 6.81LLHH189 pKa = 6.33QVFLGSLSCRR199 pKa = 11.84SVGKK203 pKa = 9.79LAQKK207 pKa = 10.43LL208 pKa = 3.71
Molecular weight: 23.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
21003233 |
17 |
7663 |
595.3 |
66.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.49 ± 0.012 | 2.257 ± 0.01 |
5.178 ± 0.009 | 6.832 ± 0.016 |
3.698 ± 0.009 | 6.318 ± 0.015 |
2.609 ± 0.007 | 4.383 ± 0.01 |
5.713 ± 0.015 | 9.486 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.326 ± 0.005 | 3.909 ± 0.009 |
5.772 ± 0.019 | 4.716 ± 0.012 |
5.652 ± 0.011 | 8.914 ± 0.018 |
5.633 ± 0.014 | 6.275 ± 0.012 |
1.144 ± 0.004 | 2.694 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
