
Tortoise microvirus 106
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
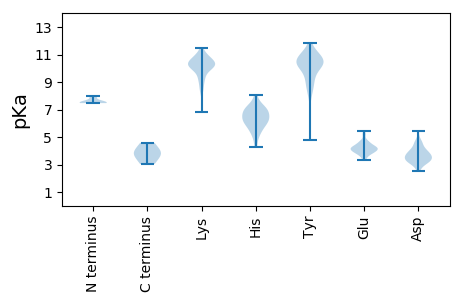
Average proteome isoelectric point is 7.38
Get precalculated fractions of proteins
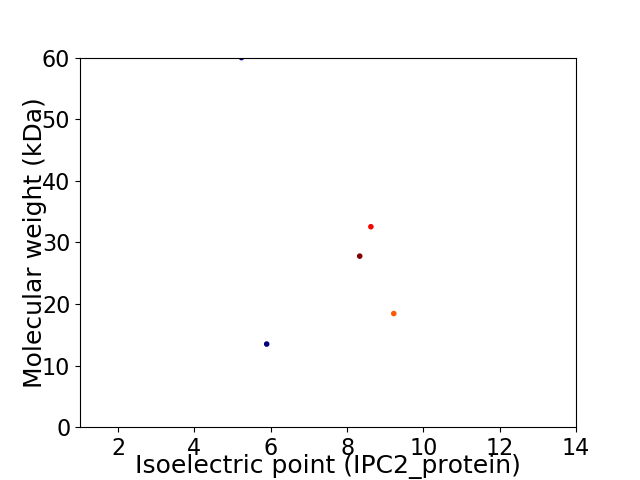
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W705|A0A4P8W705_9VIRU DNA pilot protein OS=Tortoise microvirus 106 OX=2583106 PE=4 SV=1
MM1 pKa = 7.55NSNKK5 pKa = 9.44TKK7 pKa = 10.99GNIFQNVLSADD18 pKa = 3.81VPQSQFDD25 pKa = 3.66LSFEE29 pKa = 4.24NKK31 pKa = 8.27LTCDD35 pKa = 3.43MGEE38 pKa = 5.22LIPICCQEE46 pKa = 4.5ILPGDD51 pKa = 4.08KK52 pKa = 10.17FQCDD56 pKa = 3.31ANVFIRR62 pKa = 11.84FQPLIAPIMHH72 pKa = 7.01NIDD75 pKa = 3.06VHH77 pKa = 5.64VRR79 pKa = 11.84WFFVPNRR86 pKa = 11.84LVWNEE91 pKa = 3.75FEE93 pKa = 5.42DD94 pKa = 5.42FITGGTTGMEE104 pKa = 4.21SPIYY108 pKa = 9.66PYY110 pKa = 10.46RR111 pKa = 11.84EE112 pKa = 4.06YY113 pKa = 11.06KK114 pKa = 10.28EE115 pKa = 4.17SARR118 pKa = 11.84LGSLEE123 pKa = 5.27DD124 pKa = 3.73YY125 pKa = 10.88LGVPTEE131 pKa = 4.25TPEE134 pKa = 4.73GLFGMHH140 pKa = 6.89NIIVNMLPFRR150 pKa = 11.84GYY152 pKa = 10.25ALIWNEE158 pKa = 3.64YY159 pKa = 10.36FRR161 pKa = 11.84DD162 pKa = 3.68QTLDD166 pKa = 3.12TEE168 pKa = 4.44MEE170 pKa = 3.98IARR173 pKa = 11.84GSGEE177 pKa = 3.87DD178 pKa = 3.17TKK180 pKa = 11.32DD181 pKa = 3.18YY182 pKa = 10.74FLRR185 pKa = 11.84RR186 pKa = 11.84VRR188 pKa = 11.84WEE190 pKa = 3.41KK191 pKa = 11.2DD192 pKa = 3.24YY193 pKa = 9.28FTSALPWTQRR203 pKa = 11.84GEE205 pKa = 4.2PVHH208 pKa = 6.08MPPMTLRR215 pKa = 11.84EE216 pKa = 4.14GLHH219 pKa = 6.85PDD221 pKa = 3.32MVIQDD226 pKa = 4.27TIRR229 pKa = 11.84IVDD232 pKa = 3.47ASGNTFSYY240 pKa = 10.65PEE242 pKa = 3.94NPLYY246 pKa = 10.83GNPYY250 pKa = 10.17RR251 pKa = 11.84LGHH254 pKa = 5.45QSGVVTDD261 pKa = 4.65PNEE264 pKa = 3.84KK265 pKa = 10.33PIFFDD270 pKa = 4.71FSAYY274 pKa = 10.31DD275 pKa = 3.37SYY277 pKa = 11.6TINQLRR283 pKa = 11.84EE284 pKa = 3.84ANAVQRR290 pKa = 11.84FLEE293 pKa = 4.33RR294 pKa = 11.84QAVGGARR301 pKa = 11.84YY302 pKa = 10.24AEE304 pKa = 3.99TLLNHH309 pKa = 6.81FGVRR313 pKa = 11.84TADD316 pKa = 3.31SRR318 pKa = 11.84LQRR321 pKa = 11.84PEE323 pKa = 3.5YY324 pKa = 10.45LGGHH328 pKa = 5.73RR329 pKa = 11.84QPVTVSEE336 pKa = 4.37VLQQSEE342 pKa = 4.63SSATSPQGNMAGRR355 pKa = 11.84AISTGSFGSEE365 pKa = 3.38STLFTEE371 pKa = 4.56HH372 pKa = 7.02GYY374 pKa = 11.06LFGLMYY380 pKa = 10.47VRR382 pKa = 11.84PQTGYY387 pKa = 8.53TNTFKK392 pKa = 11.23KK393 pKa = 9.75MLHH396 pKa = 5.27QKK398 pKa = 10.28ADD400 pKa = 3.34KK401 pKa = 10.35FDD403 pKa = 4.04YY404 pKa = 10.38AWPEE408 pKa = 3.78FASLGEE414 pKa = 3.93QEE416 pKa = 4.49ILQKK420 pKa = 10.13EE421 pKa = 4.48VHH423 pKa = 5.86VGTRR427 pKa = 11.84TGLEE431 pKa = 4.07SNNVFGYY438 pKa = 7.5QSRR441 pKa = 11.84YY442 pKa = 10.44AEE444 pKa = 3.83YY445 pKa = 10.54KK446 pKa = 9.1FNNNEE451 pKa = 3.63VHH453 pKa = 7.21GSFKK457 pKa = 10.61KK458 pKa = 10.55DD459 pKa = 3.03LSFWHH464 pKa = 7.13LARR467 pKa = 11.84LYY469 pKa = 10.86FRR471 pKa = 11.84DD472 pKa = 3.81YY473 pKa = 11.55DD474 pKa = 3.87PALDD478 pKa = 3.66ATFVACEE485 pKa = 3.74PRR487 pKa = 11.84KK488 pKa = 10.6DD489 pKa = 3.1IFAVVSEE496 pKa = 4.63DD497 pKa = 3.94RR498 pKa = 11.84NDD500 pKa = 3.52SLLVEE505 pKa = 4.72VYY507 pKa = 11.04NKK509 pKa = 8.52ITAIRR514 pKa = 11.84PLPAFGKK521 pKa = 9.92PSLL524 pKa = 4.01
MM1 pKa = 7.55NSNKK5 pKa = 9.44TKK7 pKa = 10.99GNIFQNVLSADD18 pKa = 3.81VPQSQFDD25 pKa = 3.66LSFEE29 pKa = 4.24NKK31 pKa = 8.27LTCDD35 pKa = 3.43MGEE38 pKa = 5.22LIPICCQEE46 pKa = 4.5ILPGDD51 pKa = 4.08KK52 pKa = 10.17FQCDD56 pKa = 3.31ANVFIRR62 pKa = 11.84FQPLIAPIMHH72 pKa = 7.01NIDD75 pKa = 3.06VHH77 pKa = 5.64VRR79 pKa = 11.84WFFVPNRR86 pKa = 11.84LVWNEE91 pKa = 3.75FEE93 pKa = 5.42DD94 pKa = 5.42FITGGTTGMEE104 pKa = 4.21SPIYY108 pKa = 9.66PYY110 pKa = 10.46RR111 pKa = 11.84EE112 pKa = 4.06YY113 pKa = 11.06KK114 pKa = 10.28EE115 pKa = 4.17SARR118 pKa = 11.84LGSLEE123 pKa = 5.27DD124 pKa = 3.73YY125 pKa = 10.88LGVPTEE131 pKa = 4.25TPEE134 pKa = 4.73GLFGMHH140 pKa = 6.89NIIVNMLPFRR150 pKa = 11.84GYY152 pKa = 10.25ALIWNEE158 pKa = 3.64YY159 pKa = 10.36FRR161 pKa = 11.84DD162 pKa = 3.68QTLDD166 pKa = 3.12TEE168 pKa = 4.44MEE170 pKa = 3.98IARR173 pKa = 11.84GSGEE177 pKa = 3.87DD178 pKa = 3.17TKK180 pKa = 11.32DD181 pKa = 3.18YY182 pKa = 10.74FLRR185 pKa = 11.84RR186 pKa = 11.84VRR188 pKa = 11.84WEE190 pKa = 3.41KK191 pKa = 11.2DD192 pKa = 3.24YY193 pKa = 9.28FTSALPWTQRR203 pKa = 11.84GEE205 pKa = 4.2PVHH208 pKa = 6.08MPPMTLRR215 pKa = 11.84EE216 pKa = 4.14GLHH219 pKa = 6.85PDD221 pKa = 3.32MVIQDD226 pKa = 4.27TIRR229 pKa = 11.84IVDD232 pKa = 3.47ASGNTFSYY240 pKa = 10.65PEE242 pKa = 3.94NPLYY246 pKa = 10.83GNPYY250 pKa = 10.17RR251 pKa = 11.84LGHH254 pKa = 5.45QSGVVTDD261 pKa = 4.65PNEE264 pKa = 3.84KK265 pKa = 10.33PIFFDD270 pKa = 4.71FSAYY274 pKa = 10.31DD275 pKa = 3.37SYY277 pKa = 11.6TINQLRR283 pKa = 11.84EE284 pKa = 3.84ANAVQRR290 pKa = 11.84FLEE293 pKa = 4.33RR294 pKa = 11.84QAVGGARR301 pKa = 11.84YY302 pKa = 10.24AEE304 pKa = 3.99TLLNHH309 pKa = 6.81FGVRR313 pKa = 11.84TADD316 pKa = 3.31SRR318 pKa = 11.84LQRR321 pKa = 11.84PEE323 pKa = 3.5YY324 pKa = 10.45LGGHH328 pKa = 5.73RR329 pKa = 11.84QPVTVSEE336 pKa = 4.37VLQQSEE342 pKa = 4.63SSATSPQGNMAGRR355 pKa = 11.84AISTGSFGSEE365 pKa = 3.38STLFTEE371 pKa = 4.56HH372 pKa = 7.02GYY374 pKa = 11.06LFGLMYY380 pKa = 10.47VRR382 pKa = 11.84PQTGYY387 pKa = 8.53TNTFKK392 pKa = 11.23KK393 pKa = 9.75MLHH396 pKa = 5.27QKK398 pKa = 10.28ADD400 pKa = 3.34KK401 pKa = 10.35FDD403 pKa = 4.04YY404 pKa = 10.38AWPEE408 pKa = 3.78FASLGEE414 pKa = 3.93QEE416 pKa = 4.49ILQKK420 pKa = 10.13EE421 pKa = 4.48VHH423 pKa = 5.86VGTRR427 pKa = 11.84TGLEE431 pKa = 4.07SNNVFGYY438 pKa = 7.5QSRR441 pKa = 11.84YY442 pKa = 10.44AEE444 pKa = 3.83YY445 pKa = 10.54KK446 pKa = 9.1FNNNEE451 pKa = 3.63VHH453 pKa = 7.21GSFKK457 pKa = 10.61KK458 pKa = 10.55DD459 pKa = 3.03LSFWHH464 pKa = 7.13LARR467 pKa = 11.84LYY469 pKa = 10.86FRR471 pKa = 11.84DD472 pKa = 3.81YY473 pKa = 11.55DD474 pKa = 3.87PALDD478 pKa = 3.66ATFVACEE485 pKa = 3.74PRR487 pKa = 11.84KK488 pKa = 10.6DD489 pKa = 3.1IFAVVSEE496 pKa = 4.63DD497 pKa = 3.94RR498 pKa = 11.84NDD500 pKa = 3.52SLLVEE505 pKa = 4.72VYY507 pKa = 11.04NKK509 pKa = 8.52ITAIRR514 pKa = 11.84PLPAFGKK521 pKa = 9.92PSLL524 pKa = 4.01
Molecular weight: 60.02 kDa
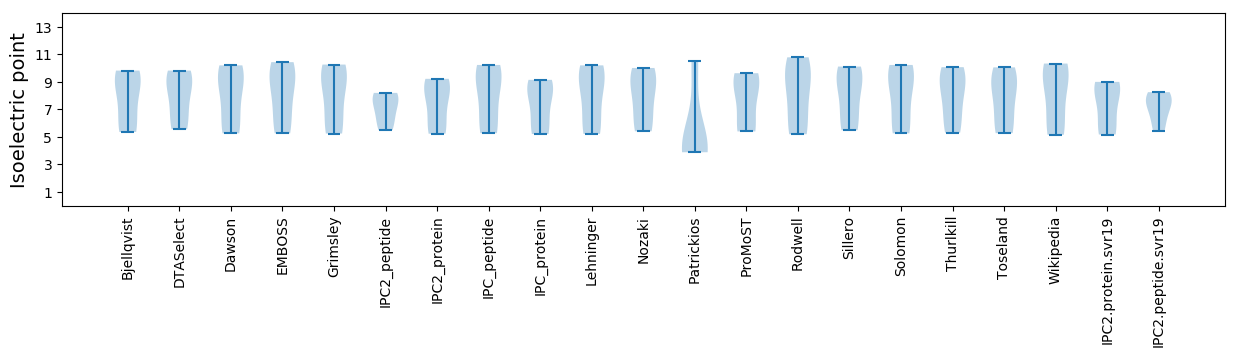
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8WAH3|A0A4P8WAH3_9VIRU Uncharacterized protein OS=Tortoise microvirus 106 OX=2583106 PE=4 SV=1
MM1 pKa = 7.53LNIEE5 pKa = 5.16KK6 pKa = 10.67SLTTNNQYY14 pKa = 10.62IIFVHH19 pKa = 6.17SYY21 pKa = 9.66KK22 pKa = 10.68NYY24 pKa = 10.3TIMEE28 pKa = 4.31TLKK31 pKa = 11.04NLLNRR36 pKa = 11.84NDD38 pKa = 3.55VVRR41 pKa = 11.84LMKK44 pKa = 10.87NLDD47 pKa = 3.73GLSLPFLFFVKK58 pKa = 10.47PSDD61 pKa = 3.67RR62 pKa = 11.84SSRR65 pKa = 11.84WLLKK69 pKa = 9.39ASKK72 pKa = 10.27RR73 pKa = 11.84IGAFGSIKK81 pKa = 10.29DD82 pKa = 3.58ISEE85 pKa = 4.72SIPILDD91 pKa = 4.64LFTGKK96 pKa = 9.73IMAITVLVRR105 pKa = 11.84DD106 pKa = 3.47EE107 pKa = 4.11WFEE110 pKa = 4.26LFVMDD115 pKa = 4.99VPFSIKK121 pKa = 9.99KK122 pKa = 9.8EE123 pKa = 3.92KK124 pKa = 10.74LLLSLFLKK132 pKa = 10.6EE133 pKa = 4.05SGNRR137 pKa = 11.84RR138 pKa = 11.84SFSYY142 pKa = 11.11NNYY145 pKa = 9.43ILQLFNSLKK154 pKa = 10.71NKK156 pKa = 10.29SLL158 pKa = 3.56
MM1 pKa = 7.53LNIEE5 pKa = 5.16KK6 pKa = 10.67SLTTNNQYY14 pKa = 10.62IIFVHH19 pKa = 6.17SYY21 pKa = 9.66KK22 pKa = 10.68NYY24 pKa = 10.3TIMEE28 pKa = 4.31TLKK31 pKa = 11.04NLLNRR36 pKa = 11.84NDD38 pKa = 3.55VVRR41 pKa = 11.84LMKK44 pKa = 10.87NLDD47 pKa = 3.73GLSLPFLFFVKK58 pKa = 10.47PSDD61 pKa = 3.67RR62 pKa = 11.84SSRR65 pKa = 11.84WLLKK69 pKa = 9.39ASKK72 pKa = 10.27RR73 pKa = 11.84IGAFGSIKK81 pKa = 10.29DD82 pKa = 3.58ISEE85 pKa = 4.72SIPILDD91 pKa = 4.64LFTGKK96 pKa = 9.73IMAITVLVRR105 pKa = 11.84DD106 pKa = 3.47EE107 pKa = 4.11WFEE110 pKa = 4.26LFVMDD115 pKa = 4.99VPFSIKK121 pKa = 9.99KK122 pKa = 9.8EE123 pKa = 3.92KK124 pKa = 10.74LLLSLFLKK132 pKa = 10.6EE133 pKa = 4.05SGNRR137 pKa = 11.84RR138 pKa = 11.84SFSYY142 pKa = 11.11NNYY145 pKa = 9.43ILQLFNSLKK154 pKa = 10.71NKK156 pKa = 10.29SLL158 pKa = 3.56
Molecular weight: 18.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1316 |
119 |
524 |
263.2 |
30.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.015 ± 0.774 | 1.14 ± 0.489 |
5.015 ± 0.401 | 6.687 ± 0.401 |
4.635 ± 1.209 | 5.167 ± 0.937 |
2.052 ± 0.554 | 5.243 ± 0.698 |
6.535 ± 1.327 | 9.574 ± 1.304 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.888 ± 0.227 | 6.687 ± 1.009 |
5.167 ± 0.722 | 4.635 ± 0.904 |
5.775 ± 0.232 | 7.447 ± 0.953 |
4.787 ± 0.921 | 4.787 ± 0.655 |
1.292 ± 0.186 | 5.471 ± 0.798 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
