
Natribacillus halophilus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Natribacillus
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
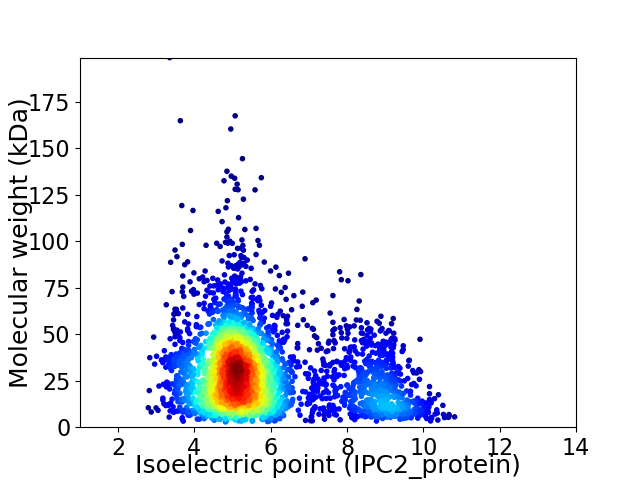
Virtual 2D-PAGE plot for 3434 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
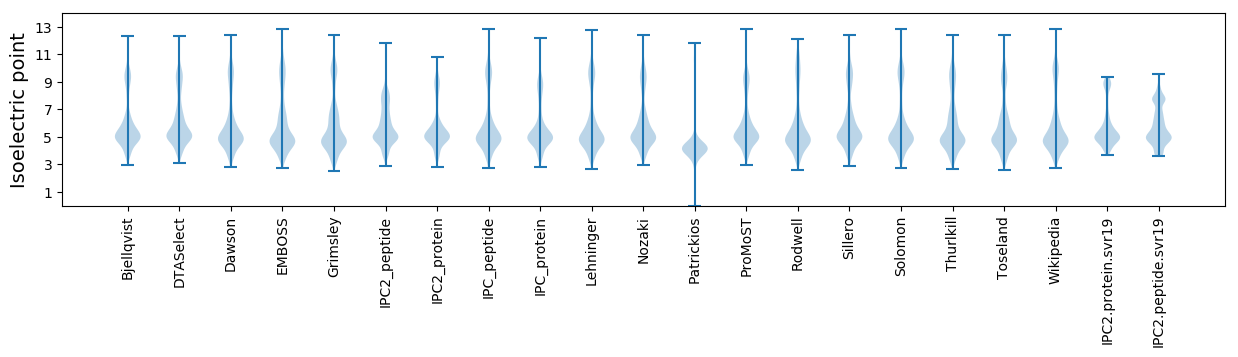
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G8RM68|A0A1G8RM68_9BACI Multimeric flavodoxin WrbA OS=Natribacillus halophilus OX=549003 GN=SAMN04488123_1197 PE=4 SV=1
MM1 pKa = 7.5SNGKK5 pKa = 7.56TRR7 pKa = 11.84MLFLSSSLVTAVMFTPMWSEE27 pKa = 3.87NVSAYY32 pKa = 10.44APDD35 pKa = 4.0EE36 pKa = 4.82DD37 pKa = 5.49DD38 pKa = 6.0PNAQDD43 pKa = 3.64YY44 pKa = 9.73TEE46 pKa = 4.83EE47 pKa = 5.4DD48 pKa = 3.42MDD50 pKa = 4.07NSQPYY55 pKa = 9.39LLKK58 pKa = 10.17HH59 pKa = 6.04DD60 pKa = 3.88QGSDD64 pKa = 3.45VEE66 pKa = 4.48QLQSDD71 pKa = 4.04LGDD74 pKa = 3.05RR75 pKa = 11.84GYY77 pKa = 11.25NVQVDD82 pKa = 5.04GIFGPEE88 pKa = 3.6TEE90 pKa = 4.61NAVKK94 pKa = 10.17AYY96 pKa = 10.47QSDD99 pKa = 3.3QGLDD103 pKa = 3.3RR104 pKa = 11.84DD105 pKa = 4.42GVAGSNTYY113 pKa = 9.53QALASDD119 pKa = 4.05EE120 pKa = 4.41TLDD123 pKa = 4.63EE124 pKa = 4.71EE125 pKa = 5.4PDD127 pKa = 3.69TATDD131 pKa = 3.63EE132 pKa = 5.18APDD135 pKa = 3.51TSRR138 pKa = 11.84NGEE141 pKa = 3.95LSNEE145 pKa = 3.87NLEE148 pKa = 4.45NQDD151 pKa = 3.27SSASEE156 pKa = 3.67IAATAEE162 pKa = 4.4SVLGTPYY169 pKa = 10.59VWGGTTPDD177 pKa = 4.03GFDD180 pKa = 2.88SSGFINYY187 pKa = 8.83VFDD190 pKa = 3.67QNGIDD195 pKa = 3.68VSRR198 pKa = 11.84THH200 pKa = 8.63AEE202 pKa = 3.24MWEE205 pKa = 3.93NDD207 pKa = 3.91GEE209 pKa = 4.8HH210 pKa = 5.39VTSMSIGDD218 pKa = 3.48VVFFEE223 pKa = 4.49GTYY226 pKa = 9.0DD227 pKa = 3.37TTGASHH233 pKa = 6.31SGIYY237 pKa = 10.04IGDD240 pKa = 3.51NQMIHH245 pKa = 6.63AGSEE249 pKa = 4.29GVVQADD255 pKa = 3.21ITSDD259 pKa = 2.79YY260 pKa = 9.4WQDD263 pKa = 2.94HH264 pKa = 6.33FIGFKK269 pKa = 10.12TMQQ272 pKa = 3.35
MM1 pKa = 7.5SNGKK5 pKa = 7.56TRR7 pKa = 11.84MLFLSSSLVTAVMFTPMWSEE27 pKa = 3.87NVSAYY32 pKa = 10.44APDD35 pKa = 4.0EE36 pKa = 4.82DD37 pKa = 5.49DD38 pKa = 6.0PNAQDD43 pKa = 3.64YY44 pKa = 9.73TEE46 pKa = 4.83EE47 pKa = 5.4DD48 pKa = 3.42MDD50 pKa = 4.07NSQPYY55 pKa = 9.39LLKK58 pKa = 10.17HH59 pKa = 6.04DD60 pKa = 3.88QGSDD64 pKa = 3.45VEE66 pKa = 4.48QLQSDD71 pKa = 4.04LGDD74 pKa = 3.05RR75 pKa = 11.84GYY77 pKa = 11.25NVQVDD82 pKa = 5.04GIFGPEE88 pKa = 3.6TEE90 pKa = 4.61NAVKK94 pKa = 10.17AYY96 pKa = 10.47QSDD99 pKa = 3.3QGLDD103 pKa = 3.3RR104 pKa = 11.84DD105 pKa = 4.42GVAGSNTYY113 pKa = 9.53QALASDD119 pKa = 4.05EE120 pKa = 4.41TLDD123 pKa = 4.63EE124 pKa = 4.71EE125 pKa = 5.4PDD127 pKa = 3.69TATDD131 pKa = 3.63EE132 pKa = 5.18APDD135 pKa = 3.51TSRR138 pKa = 11.84NGEE141 pKa = 3.95LSNEE145 pKa = 3.87NLEE148 pKa = 4.45NQDD151 pKa = 3.27SSASEE156 pKa = 3.67IAATAEE162 pKa = 4.4SVLGTPYY169 pKa = 10.59VWGGTTPDD177 pKa = 4.03GFDD180 pKa = 2.88SSGFINYY187 pKa = 8.83VFDD190 pKa = 3.67QNGIDD195 pKa = 3.68VSRR198 pKa = 11.84THH200 pKa = 8.63AEE202 pKa = 3.24MWEE205 pKa = 3.93NDD207 pKa = 3.91GEE209 pKa = 4.8HH210 pKa = 5.39VTSMSIGDD218 pKa = 3.48VVFFEE223 pKa = 4.49GTYY226 pKa = 9.0DD227 pKa = 3.37TTGASHH233 pKa = 6.31SGIYY237 pKa = 10.04IGDD240 pKa = 3.51NQMIHH245 pKa = 6.63AGSEE249 pKa = 4.29GVVQADD255 pKa = 3.21ITSDD259 pKa = 2.79YY260 pKa = 9.4WQDD263 pKa = 2.94HH264 pKa = 6.33FIGFKK269 pKa = 10.12TMQQ272 pKa = 3.35
Molecular weight: 29.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G8KHB6|A0A1G8KHB6_9BACI Uncharacterized protein OS=Natribacillus halophilus OX=549003 GN=SAMN04488123_102102 PE=4 SV=1
MM1 pKa = 7.23TLEE4 pKa = 4.3SCCYY8 pKa = 8.23TPKK11 pKa = 10.61AGYY14 pKa = 8.7LAPRR18 pKa = 11.84GTRR21 pKa = 11.84SALLYY26 pKa = 8.53PKK28 pKa = 10.37SRR30 pKa = 11.84VSHH33 pKa = 5.16SRR35 pKa = 11.84RR36 pKa = 11.84TRR38 pKa = 11.84SALLYY43 pKa = 8.47PKK45 pKa = 10.45GRR47 pKa = 11.84VSHH50 pKa = 5.85SEE52 pKa = 3.62RR53 pKa = 11.84DD54 pKa = 3.45KK55 pKa = 11.28KK56 pKa = 10.77RR57 pKa = 11.84VVIPQRR63 pKa = 11.84QGISLRR69 pKa = 11.84EE70 pKa = 4.01GPEE73 pKa = 3.88ACCYY77 pKa = 8.47TPKK80 pKa = 10.62AGYY83 pKa = 7.53LTPRR87 pKa = 11.84GTRR90 pKa = 11.84SALLYY95 pKa = 9.27PKK97 pKa = 10.34SRR99 pKa = 11.84VSRR102 pKa = 11.84SEE104 pKa = 3.61RR105 pKa = 11.84DD106 pKa = 3.41KK107 pKa = 11.45KK108 pKa = 10.58RR109 pKa = 11.84VAIPQKK115 pKa = 10.21QGISLRR121 pKa = 11.84EE122 pKa = 3.94GQEE125 pKa = 3.95ACCYY129 pKa = 8.15TPKK132 pKa = 10.55AGYY135 pKa = 7.19PAPRR139 pKa = 11.84GTKK142 pKa = 8.62NHH144 pKa = 6.28VIIPRR149 pKa = 11.84TTGIEE154 pKa = 4.22PIRR157 pKa = 11.84EE158 pKa = 3.86GDD160 pKa = 3.73VILPRR165 pKa = 11.84ARR167 pKa = 11.84LRR169 pKa = 11.84KK170 pKa = 9.86AIFISLKK177 pKa = 10.42NRR179 pKa = 11.84FTLTLPWPISSFCCDD194 pKa = 2.79WHH196 pKa = 7.59SEE198 pKa = 4.17PLQSTT203 pKa = 3.96
MM1 pKa = 7.23TLEE4 pKa = 4.3SCCYY8 pKa = 8.23TPKK11 pKa = 10.61AGYY14 pKa = 8.7LAPRR18 pKa = 11.84GTRR21 pKa = 11.84SALLYY26 pKa = 8.53PKK28 pKa = 10.37SRR30 pKa = 11.84VSHH33 pKa = 5.16SRR35 pKa = 11.84RR36 pKa = 11.84TRR38 pKa = 11.84SALLYY43 pKa = 8.47PKK45 pKa = 10.45GRR47 pKa = 11.84VSHH50 pKa = 5.85SEE52 pKa = 3.62RR53 pKa = 11.84DD54 pKa = 3.45KK55 pKa = 11.28KK56 pKa = 10.77RR57 pKa = 11.84VVIPQRR63 pKa = 11.84QGISLRR69 pKa = 11.84EE70 pKa = 4.01GPEE73 pKa = 3.88ACCYY77 pKa = 8.47TPKK80 pKa = 10.62AGYY83 pKa = 7.53LTPRR87 pKa = 11.84GTRR90 pKa = 11.84SALLYY95 pKa = 9.27PKK97 pKa = 10.34SRR99 pKa = 11.84VSRR102 pKa = 11.84SEE104 pKa = 3.61RR105 pKa = 11.84DD106 pKa = 3.41KK107 pKa = 11.45KK108 pKa = 10.58RR109 pKa = 11.84VAIPQKK115 pKa = 10.21QGISLRR121 pKa = 11.84EE122 pKa = 3.94GQEE125 pKa = 3.95ACCYY129 pKa = 8.15TPKK132 pKa = 10.55AGYY135 pKa = 7.19PAPRR139 pKa = 11.84GTKK142 pKa = 8.62NHH144 pKa = 6.28VIIPRR149 pKa = 11.84TTGIEE154 pKa = 4.22PIRR157 pKa = 11.84EE158 pKa = 3.86GDD160 pKa = 3.73VILPRR165 pKa = 11.84ARR167 pKa = 11.84LRR169 pKa = 11.84KK170 pKa = 9.86AIFISLKK177 pKa = 10.42NRR179 pKa = 11.84FTLTLPWPISSFCCDD194 pKa = 2.79WHH196 pKa = 7.59SEE198 pKa = 4.17PLQSTT203 pKa = 3.96
Molecular weight: 22.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
952482 |
27 |
1818 |
277.4 |
31.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.008 ± 0.041 | 0.641 ± 0.011 |
6.008 ± 0.042 | 8.273 ± 0.062 |
4.17 ± 0.035 | 7.24 ± 0.04 |
2.38 ± 0.023 | 6.812 ± 0.042 |
4.972 ± 0.045 | 9.373 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.003 ± 0.022 | 3.807 ± 0.027 |
3.86 ± 0.024 | 3.961 ± 0.031 |
4.768 ± 0.036 | 5.597 ± 0.028 |
5.595 ± 0.026 | 7.207 ± 0.033 |
1.047 ± 0.014 | 3.276 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
