
Serinus canaria papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Etapapillomavirus; unclassified Etapapillomavirus
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
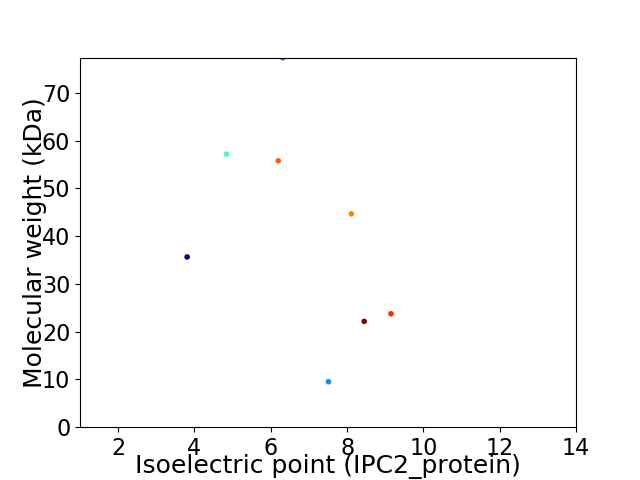
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L2P5B7|A0A2L2P5B7_9PAPI L2 OS=Serinus canaria papillomavirus 1 OX=2094713 PE=4 SV=1
MM1 pKa = 7.69PRR3 pKa = 11.84QPLWSPPVRR12 pKa = 11.84QPEE15 pKa = 4.23EE16 pKa = 3.84NDD18 pKa = 2.85EE19 pKa = 5.59DD20 pKa = 4.05IDD22 pKa = 4.25FNLFIDD28 pKa = 4.33DD29 pKa = 4.58SEE31 pKa = 4.82NNSSDD36 pKa = 4.76SEE38 pKa = 4.53GSWEE42 pKa = 4.55LCLTVQGSEE51 pKa = 4.12HH52 pKa = 6.66VFDD55 pKa = 5.02GLSEE59 pKa = 4.07DD60 pKa = 4.02VEE62 pKa = 4.74GGEE65 pKa = 4.25SDD67 pKa = 5.34SYY69 pKa = 11.3IQGLDD74 pKa = 3.53VPDD77 pKa = 4.94GVCPSLVCTEE87 pKa = 4.78DD88 pKa = 3.55PSFSSPEE95 pKa = 4.12LGIHH99 pKa = 6.67PALTPGLFTTAPEE112 pKa = 4.4VVPPPYY118 pKa = 9.88RR119 pKa = 11.84PPPPYY124 pKa = 9.84PGLPPPYY131 pKa = 9.32RR132 pKa = 11.84PQPPPCRR139 pKa = 11.84PPPPPCRR146 pKa = 11.84PPPPPYY152 pKa = 10.04RR153 pKa = 11.84PPPPPYY159 pKa = 9.9RR160 pKa = 11.84PPPPPYY166 pKa = 9.94RR167 pKa = 11.84PPACPTPAYY176 pKa = 9.45PEE178 pKa = 4.7LPACRR183 pKa = 11.84YY184 pKa = 5.58TAPGIGSAYY193 pKa = 8.91GQNPGPYY200 pKa = 8.5GQNPSYY206 pKa = 10.13EE207 pKa = 4.12PSYY210 pKa = 10.28EE211 pKa = 3.85PSYY214 pKa = 10.46QPSYY218 pKa = 9.79QPSYY222 pKa = 9.8QPSYY226 pKa = 9.97QQDD229 pKa = 3.62PYY231 pKa = 10.76YY232 pKa = 10.52GQNPGHH238 pKa = 6.07GQDD241 pKa = 3.82PASGQLPNGPPASAPYY257 pKa = 9.74YY258 pKa = 10.22PPDD261 pKa = 3.7TLQVGLDD268 pKa = 3.65VEE270 pKa = 4.45DD271 pKa = 4.57QEE273 pKa = 5.79DD274 pKa = 4.55GPGYY278 pKa = 10.36SISTNWACTICGNPLTLTDD297 pKa = 4.24LLMYY301 pKa = 10.15EE302 pKa = 4.87PVSQAQQLGVCYY314 pKa = 10.24LCYY317 pKa = 10.24SGQSLEE323 pKa = 5.17DD324 pKa = 3.6LFASS328 pKa = 4.24
MM1 pKa = 7.69PRR3 pKa = 11.84QPLWSPPVRR12 pKa = 11.84QPEE15 pKa = 4.23EE16 pKa = 3.84NDD18 pKa = 2.85EE19 pKa = 5.59DD20 pKa = 4.05IDD22 pKa = 4.25FNLFIDD28 pKa = 4.33DD29 pKa = 4.58SEE31 pKa = 4.82NNSSDD36 pKa = 4.76SEE38 pKa = 4.53GSWEE42 pKa = 4.55LCLTVQGSEE51 pKa = 4.12HH52 pKa = 6.66VFDD55 pKa = 5.02GLSEE59 pKa = 4.07DD60 pKa = 4.02VEE62 pKa = 4.74GGEE65 pKa = 4.25SDD67 pKa = 5.34SYY69 pKa = 11.3IQGLDD74 pKa = 3.53VPDD77 pKa = 4.94GVCPSLVCTEE87 pKa = 4.78DD88 pKa = 3.55PSFSSPEE95 pKa = 4.12LGIHH99 pKa = 6.67PALTPGLFTTAPEE112 pKa = 4.4VVPPPYY118 pKa = 9.88RR119 pKa = 11.84PPPPYY124 pKa = 9.84PGLPPPYY131 pKa = 9.32RR132 pKa = 11.84PQPPPCRR139 pKa = 11.84PPPPPCRR146 pKa = 11.84PPPPPYY152 pKa = 10.04RR153 pKa = 11.84PPPPPYY159 pKa = 9.9RR160 pKa = 11.84PPPPPYY166 pKa = 9.94RR167 pKa = 11.84PPACPTPAYY176 pKa = 9.45PEE178 pKa = 4.7LPACRR183 pKa = 11.84YY184 pKa = 5.58TAPGIGSAYY193 pKa = 8.91GQNPGPYY200 pKa = 8.5GQNPSYY206 pKa = 10.13EE207 pKa = 4.12PSYY210 pKa = 10.28EE211 pKa = 3.85PSYY214 pKa = 10.46QPSYY218 pKa = 9.79QPSYY222 pKa = 9.8QPSYY226 pKa = 9.97QQDD229 pKa = 3.62PYY231 pKa = 10.76YY232 pKa = 10.52GQNPGHH238 pKa = 6.07GQDD241 pKa = 3.82PASGQLPNGPPASAPYY257 pKa = 9.74YY258 pKa = 10.22PPDD261 pKa = 3.7TLQVGLDD268 pKa = 3.65VEE270 pKa = 4.45DD271 pKa = 4.57QEE273 pKa = 5.79DD274 pKa = 4.55GPGYY278 pKa = 10.36SISTNWACTICGNPLTLTDD297 pKa = 4.24LLMYY301 pKa = 10.15EE302 pKa = 4.87PVSQAQQLGVCYY314 pKa = 10.24LCYY317 pKa = 10.24SGQSLEE323 pKa = 5.17DD324 pKa = 3.6LFASS328 pKa = 4.24
Molecular weight: 35.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L2P5B0|A0A2L2P5B0_9PAPI Major capsid protein L1 OS=Serinus canaria papillomavirus 1 OX=2094713 GN=L1 PE=3 SV=1
MM1 pKa = 7.58PMWTVLGLSDD11 pKa = 4.37EE12 pKa = 4.78VPDD15 pKa = 3.82EE16 pKa = 4.42VPVPVHH22 pKa = 5.86EE23 pKa = 4.43STRR26 pKa = 11.84EE27 pKa = 3.99DD28 pKa = 3.54EE29 pKa = 4.4VDD31 pKa = 3.25RR32 pKa = 11.84SLVSTPPPPDD42 pKa = 3.26LSRR45 pKa = 11.84YY46 pKa = 7.99SPRR49 pKa = 11.84PSAPRR54 pKa = 11.84KK55 pKa = 9.16GKK57 pKa = 7.3GTRR60 pKa = 11.84VVRR63 pKa = 11.84PTRR66 pKa = 11.84PGPSGRR72 pKa = 11.84SSTQATQPRR81 pKa = 11.84QPATSRR87 pKa = 11.84QSVRR91 pKa = 11.84QSRR94 pKa = 11.84EE95 pKa = 3.48GLKK98 pKa = 10.49RR99 pKa = 11.84PLQPPTPDD107 pKa = 2.72EE108 pKa = 4.36VGTVHH113 pKa = 7.01SSSHH117 pKa = 5.93HH118 pKa = 5.54GGTRR122 pKa = 11.84VDD124 pKa = 5.05RR125 pKa = 11.84LIQDD129 pKa = 3.34ARR131 pKa = 11.84DD132 pKa = 3.6PPGLCFEE139 pKa = 5.03GKK141 pKa = 8.43TNQLKK146 pKa = 9.62TIRR149 pKa = 11.84QRR151 pKa = 11.84IFGGQLPFEE160 pKa = 4.31RR161 pKa = 11.84VSTTWHH167 pKa = 5.48WVKK170 pKa = 10.97KK171 pKa = 10.45DD172 pKa = 3.44GEE174 pKa = 4.43SDD176 pKa = 3.64SKK178 pKa = 11.16ILVVFKK184 pKa = 10.66DD185 pKa = 3.15KK186 pKa = 10.78KK187 pKa = 10.98DD188 pKa = 3.23RR189 pKa = 11.84DD190 pKa = 3.81FFQKK194 pKa = 10.38SFYY197 pKa = 10.27VGSSGVRR204 pKa = 11.84VFHH207 pKa = 7.11CSLAGLL213 pKa = 4.12
MM1 pKa = 7.58PMWTVLGLSDD11 pKa = 4.37EE12 pKa = 4.78VPDD15 pKa = 3.82EE16 pKa = 4.42VPVPVHH22 pKa = 5.86EE23 pKa = 4.43STRR26 pKa = 11.84EE27 pKa = 3.99DD28 pKa = 3.54EE29 pKa = 4.4VDD31 pKa = 3.25RR32 pKa = 11.84SLVSTPPPPDD42 pKa = 3.26LSRR45 pKa = 11.84YY46 pKa = 7.99SPRR49 pKa = 11.84PSAPRR54 pKa = 11.84KK55 pKa = 9.16GKK57 pKa = 7.3GTRR60 pKa = 11.84VVRR63 pKa = 11.84PTRR66 pKa = 11.84PGPSGRR72 pKa = 11.84SSTQATQPRR81 pKa = 11.84QPATSRR87 pKa = 11.84QSVRR91 pKa = 11.84QSRR94 pKa = 11.84EE95 pKa = 3.48GLKK98 pKa = 10.49RR99 pKa = 11.84PLQPPTPDD107 pKa = 2.72EE108 pKa = 4.36VGTVHH113 pKa = 7.01SSSHH117 pKa = 5.93HH118 pKa = 5.54GGTRR122 pKa = 11.84VDD124 pKa = 5.05RR125 pKa = 11.84LIQDD129 pKa = 3.34ARR131 pKa = 11.84DD132 pKa = 3.6PPGLCFEE139 pKa = 5.03GKK141 pKa = 8.43TNQLKK146 pKa = 9.62TIRR149 pKa = 11.84QRR151 pKa = 11.84IFGGQLPFEE160 pKa = 4.31RR161 pKa = 11.84VSTTWHH167 pKa = 5.48WVKK170 pKa = 10.97KK171 pKa = 10.45DD172 pKa = 3.44GEE174 pKa = 4.43SDD176 pKa = 3.64SKK178 pKa = 11.16ILVVFKK184 pKa = 10.66DD185 pKa = 3.15KK186 pKa = 10.78KK187 pKa = 10.98DD188 pKa = 3.23RR189 pKa = 11.84DD190 pKa = 3.81FFQKK194 pKa = 10.38SFYY197 pKa = 10.27VGSSGVRR204 pKa = 11.84VFHH207 pKa = 7.11CSLAGLL213 pKa = 4.12
Molecular weight: 23.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2925 |
83 |
678 |
365.6 |
40.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.154 ± 0.635 | 2.12 ± 0.556 |
6.017 ± 0.315 | 5.709 ± 0.324 |
3.487 ± 0.36 | 7.556 ± 0.934 |
2.154 ± 0.324 | 3.897 ± 0.458 |
3.658 ± 0.777 | 8.308 ± 0.897 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.436 ± 0.226 | 3.179 ± 0.536 |
9.299 ± 1.807 | 4.821 ± 0.414 |
7.214 ± 0.679 | 8.068 ± 0.545 |
5.231 ± 0.646 | 6.359 ± 0.887 |
1.607 ± 0.132 | 3.726 ± 0.58 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
