
Capybara microvirus Cap1_SP_83
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.68
Get precalculated fractions of proteins
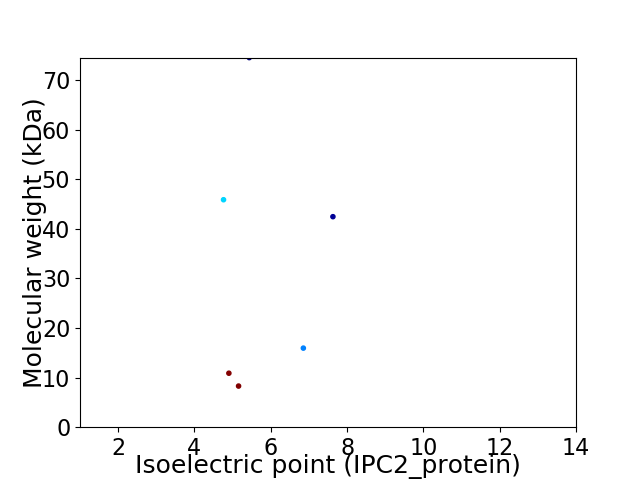
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7M1|A0A4P8W7M1_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_83 OX=2584794 PE=4 SV=1
MM1 pKa = 7.57SLLDD5 pKa = 4.44NIKK8 pKa = 9.66TSAKK12 pKa = 9.64EE13 pKa = 3.75DD14 pKa = 3.69SLLVSQSNSALLDD27 pKa = 3.74SLAHH31 pKa = 5.95QFDD34 pKa = 3.95GTDD37 pKa = 3.6FEE39 pKa = 6.36SRR41 pKa = 11.84AQSNPYY47 pKa = 8.2LTSSYY52 pKa = 10.96GSKK55 pKa = 9.06TLWDD59 pKa = 4.55KK60 pKa = 11.28IGDD63 pKa = 4.05LIGFRR68 pKa = 11.84TSEE71 pKa = 4.12DD72 pKa = 3.47VYY74 pKa = 10.64EE75 pKa = 4.19TDD77 pKa = 5.0RR78 pKa = 11.84IQAAKK83 pKa = 10.28DD84 pKa = 3.37YY85 pKa = 10.6NAQLQQLYY93 pKa = 10.76DD94 pKa = 3.69EE95 pKa = 4.6QAYY98 pKa = 10.57NSPSAQASRR107 pKa = 11.84YY108 pKa = 9.11RR109 pKa = 11.84EE110 pKa = 3.79AGINPDD116 pKa = 2.99INGSVSSGEE125 pKa = 3.68ASEE128 pKa = 5.24FSQEE132 pKa = 3.77QTSPVSPPASYY143 pKa = 11.49NFGQFSKK150 pKa = 10.98VCFDD154 pKa = 5.46AIQVALNLADD164 pKa = 3.98GFTSLRR170 pKa = 11.84GKK172 pKa = 10.7FIDD175 pKa = 3.53QAGAEE180 pKa = 4.19LSQYY184 pKa = 10.88SAVDD188 pKa = 3.61SVALLDD194 pKa = 4.66FLSQEE199 pKa = 4.38QISDD203 pKa = 3.75SDD205 pKa = 3.62FDD207 pKa = 5.45DD208 pKa = 4.56YY209 pKa = 11.87PNMIAPAVKK218 pKa = 10.3VEE220 pKa = 3.99KK221 pKa = 11.0GPDD224 pKa = 3.13KK225 pKa = 10.79FSSKK229 pKa = 10.04RR230 pKa = 11.84LQSAYY235 pKa = 10.25DD236 pKa = 3.37RR237 pKa = 11.84KK238 pKa = 10.39LSQLIRR244 pKa = 11.84SGSLRR249 pKa = 11.84GQGIRR254 pKa = 11.84MQGLNQYY261 pKa = 10.08YY262 pKa = 10.32GDD264 pKa = 3.85RR265 pKa = 11.84LSMFEE270 pKa = 4.23SKK272 pKa = 10.32NAYY275 pKa = 9.73GLDD278 pKa = 3.86GLDD281 pKa = 3.47SSLDD285 pKa = 3.24IITRR289 pKa = 11.84HH290 pKa = 5.93LGSLRR295 pKa = 11.84KK296 pKa = 8.67ATEE299 pKa = 3.66EE300 pKa = 3.97ATLKK304 pKa = 11.0ASAAQGKK311 pKa = 9.48FDD313 pKa = 3.58EE314 pKa = 6.0AYY316 pKa = 8.87TQGLDD321 pKa = 3.36PSQVSEE327 pKa = 4.78SDD329 pKa = 3.14MASYY333 pKa = 10.64KK334 pKa = 10.62SSEE337 pKa = 3.96QEE339 pKa = 3.95KK340 pKa = 10.45EE341 pKa = 3.54ISEE344 pKa = 4.28ALNKK348 pKa = 8.46TVNGIVSDD356 pKa = 3.75LRR358 pKa = 11.84KK359 pKa = 10.6NIDD362 pKa = 4.58DD363 pKa = 4.32DD364 pKa = 4.13PKK366 pKa = 10.42HH367 pKa = 6.55AFFSRR372 pKa = 11.84LLLIGFYY379 pKa = 10.86LLSNNLMPSLSFGRR393 pKa = 11.84SSSQGAYY400 pKa = 7.92TGKK403 pKa = 10.17FGSSARR409 pKa = 11.84SSSGFNFGIKK419 pKa = 9.79
MM1 pKa = 7.57SLLDD5 pKa = 4.44NIKK8 pKa = 9.66TSAKK12 pKa = 9.64EE13 pKa = 3.75DD14 pKa = 3.69SLLVSQSNSALLDD27 pKa = 3.74SLAHH31 pKa = 5.95QFDD34 pKa = 3.95GTDD37 pKa = 3.6FEE39 pKa = 6.36SRR41 pKa = 11.84AQSNPYY47 pKa = 8.2LTSSYY52 pKa = 10.96GSKK55 pKa = 9.06TLWDD59 pKa = 4.55KK60 pKa = 11.28IGDD63 pKa = 4.05LIGFRR68 pKa = 11.84TSEE71 pKa = 4.12DD72 pKa = 3.47VYY74 pKa = 10.64EE75 pKa = 4.19TDD77 pKa = 5.0RR78 pKa = 11.84IQAAKK83 pKa = 10.28DD84 pKa = 3.37YY85 pKa = 10.6NAQLQQLYY93 pKa = 10.76DD94 pKa = 3.69EE95 pKa = 4.6QAYY98 pKa = 10.57NSPSAQASRR107 pKa = 11.84YY108 pKa = 9.11RR109 pKa = 11.84EE110 pKa = 3.79AGINPDD116 pKa = 2.99INGSVSSGEE125 pKa = 3.68ASEE128 pKa = 5.24FSQEE132 pKa = 3.77QTSPVSPPASYY143 pKa = 11.49NFGQFSKK150 pKa = 10.98VCFDD154 pKa = 5.46AIQVALNLADD164 pKa = 3.98GFTSLRR170 pKa = 11.84GKK172 pKa = 10.7FIDD175 pKa = 3.53QAGAEE180 pKa = 4.19LSQYY184 pKa = 10.88SAVDD188 pKa = 3.61SVALLDD194 pKa = 4.66FLSQEE199 pKa = 4.38QISDD203 pKa = 3.75SDD205 pKa = 3.62FDD207 pKa = 5.45DD208 pKa = 4.56YY209 pKa = 11.87PNMIAPAVKK218 pKa = 10.3VEE220 pKa = 3.99KK221 pKa = 11.0GPDD224 pKa = 3.13KK225 pKa = 10.79FSSKK229 pKa = 10.04RR230 pKa = 11.84LQSAYY235 pKa = 10.25DD236 pKa = 3.37RR237 pKa = 11.84KK238 pKa = 10.39LSQLIRR244 pKa = 11.84SGSLRR249 pKa = 11.84GQGIRR254 pKa = 11.84MQGLNQYY261 pKa = 10.08YY262 pKa = 10.32GDD264 pKa = 3.85RR265 pKa = 11.84LSMFEE270 pKa = 4.23SKK272 pKa = 10.32NAYY275 pKa = 9.73GLDD278 pKa = 3.86GLDD281 pKa = 3.47SSLDD285 pKa = 3.24IITRR289 pKa = 11.84HH290 pKa = 5.93LGSLRR295 pKa = 11.84KK296 pKa = 8.67ATEE299 pKa = 3.66EE300 pKa = 3.97ATLKK304 pKa = 11.0ASAAQGKK311 pKa = 9.48FDD313 pKa = 3.58EE314 pKa = 6.0AYY316 pKa = 8.87TQGLDD321 pKa = 3.36PSQVSEE327 pKa = 4.78SDD329 pKa = 3.14MASYY333 pKa = 10.64KK334 pKa = 10.62SSEE337 pKa = 3.96QEE339 pKa = 3.95KK340 pKa = 10.45EE341 pKa = 3.54ISEE344 pKa = 4.28ALNKK348 pKa = 8.46TVNGIVSDD356 pKa = 3.75LRR358 pKa = 11.84KK359 pKa = 10.6NIDD362 pKa = 4.58DD363 pKa = 4.32DD364 pKa = 4.13PKK366 pKa = 10.42HH367 pKa = 6.55AFFSRR372 pKa = 11.84LLLIGFYY379 pKa = 10.86LLSNNLMPSLSFGRR393 pKa = 11.84SSSQGAYY400 pKa = 7.92TGKK403 pKa = 10.17FGSSARR409 pKa = 11.84SSSGFNFGIKK419 pKa = 9.79
Molecular weight: 45.86 kDa
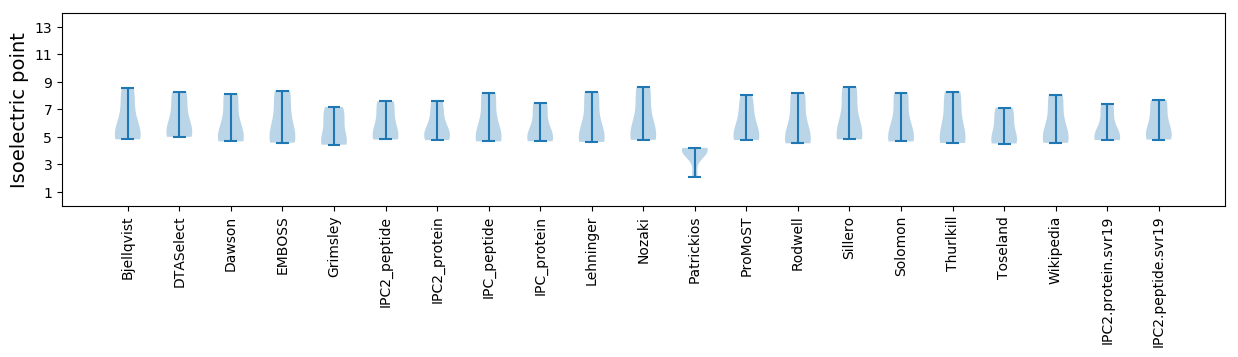
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7L4|A0A4P8W7L4_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_83 OX=2584794 PE=4 SV=1
MM1 pKa = 7.65CSSPITIRR9 pKa = 11.84NPNYY13 pKa = 10.3AFSRR17 pKa = 11.84DD18 pKa = 3.21STGNIKK24 pKa = 10.09PLIFHH29 pKa = 7.51DD30 pKa = 4.01SNIRR34 pKa = 11.84HH35 pKa = 5.74VADD38 pKa = 3.68SSPYY42 pKa = 7.76MQVPCGHH49 pKa = 6.75CPDD52 pKa = 3.7CVARR56 pKa = 11.84KK57 pKa = 10.05SSDD60 pKa = 3.01LAQRR64 pKa = 11.84ASIEE68 pKa = 4.13SLYY71 pKa = 11.1SHH73 pKa = 6.73VFFLTLTYY81 pKa = 11.28NNDD84 pKa = 3.65SLPHH88 pKa = 6.24FLTSEE93 pKa = 4.03GGNIPFADD101 pKa = 4.42FSDD104 pKa = 3.78LQKK107 pKa = 10.21MFKK110 pKa = 10.53LIRR113 pKa = 11.84KK114 pKa = 8.33YY115 pKa = 11.14NEE117 pKa = 3.21ILRR120 pKa = 11.84PFKK123 pKa = 10.51YY124 pKa = 10.33VSVSEE129 pKa = 4.44RR130 pKa = 11.84GTEE133 pKa = 4.0GARR136 pKa = 11.84PHH138 pKa = 5.73FHH140 pKa = 6.29VLLFVEE146 pKa = 5.23KK147 pKa = 10.63LPKK150 pKa = 9.97DD151 pKa = 4.02DD152 pKa = 5.36YY153 pKa = 12.05NDD155 pKa = 3.49ILNLEE160 pKa = 4.77SYY162 pKa = 10.62LSSLFLRR169 pKa = 11.84RR170 pKa = 11.84WQRR173 pKa = 11.84NYY175 pKa = 10.93GSTRR179 pKa = 11.84NPDD182 pKa = 3.31YY183 pKa = 11.28RR184 pKa = 11.84NLCTFVDD191 pKa = 3.58KK192 pKa = 10.75STLFRR197 pKa = 11.84KK198 pKa = 7.64QTNFDD203 pKa = 3.33LHH205 pKa = 5.76YY206 pKa = 10.86VKK208 pKa = 10.54PDD210 pKa = 3.32VNNGISSVVYY220 pKa = 9.58YY221 pKa = 10.42CMKK224 pKa = 10.85YY225 pKa = 8.33MLKK228 pKa = 10.37EE229 pKa = 3.99SSHH232 pKa = 6.7DD233 pKa = 3.99SKK235 pKa = 11.43LHH237 pKa = 5.26YY238 pKa = 10.03ALKK241 pKa = 10.67SSLPPGEE248 pKa = 4.94AAQSWSVVRR257 pKa = 11.84SKK259 pKa = 10.62RR260 pKa = 11.84ACSLDD265 pKa = 3.39FGRR268 pKa = 11.84KK269 pKa = 8.89SDD271 pKa = 3.65PSVLKK276 pKa = 10.51RR277 pKa = 11.84IHH279 pKa = 7.54DD280 pKa = 4.31DD281 pKa = 3.25VDD283 pKa = 3.04KK284 pKa = 11.21CRR286 pKa = 11.84YY287 pKa = 9.46LYY289 pKa = 7.42NTPVFCYY296 pKa = 9.59PDD298 pKa = 3.79CALTAPLSRR307 pKa = 11.84YY308 pKa = 10.11YY309 pKa = 11.07YY310 pKa = 10.46DD311 pKa = 5.26ALSLDD316 pKa = 3.32QVQYY320 pKa = 11.7YY321 pKa = 10.42NDD323 pKa = 4.97LYY325 pKa = 11.27QSLPPSSSLDD335 pKa = 3.38YY336 pKa = 10.89DD337 pKa = 3.61SRR339 pKa = 11.84LLKK342 pKa = 10.92DD343 pKa = 2.9SDD345 pKa = 3.17KK346 pKa = 10.71FRR348 pKa = 11.84RR349 pKa = 11.84LKK351 pKa = 10.16TDD353 pKa = 3.13VFSEE357 pKa = 4.06NVNIYY362 pKa = 10.89EE363 pKa = 4.68NILL366 pKa = 3.49
MM1 pKa = 7.65CSSPITIRR9 pKa = 11.84NPNYY13 pKa = 10.3AFSRR17 pKa = 11.84DD18 pKa = 3.21STGNIKK24 pKa = 10.09PLIFHH29 pKa = 7.51DD30 pKa = 4.01SNIRR34 pKa = 11.84HH35 pKa = 5.74VADD38 pKa = 3.68SSPYY42 pKa = 7.76MQVPCGHH49 pKa = 6.75CPDD52 pKa = 3.7CVARR56 pKa = 11.84KK57 pKa = 10.05SSDD60 pKa = 3.01LAQRR64 pKa = 11.84ASIEE68 pKa = 4.13SLYY71 pKa = 11.1SHH73 pKa = 6.73VFFLTLTYY81 pKa = 11.28NNDD84 pKa = 3.65SLPHH88 pKa = 6.24FLTSEE93 pKa = 4.03GGNIPFADD101 pKa = 4.42FSDD104 pKa = 3.78LQKK107 pKa = 10.21MFKK110 pKa = 10.53LIRR113 pKa = 11.84KK114 pKa = 8.33YY115 pKa = 11.14NEE117 pKa = 3.21ILRR120 pKa = 11.84PFKK123 pKa = 10.51YY124 pKa = 10.33VSVSEE129 pKa = 4.44RR130 pKa = 11.84GTEE133 pKa = 4.0GARR136 pKa = 11.84PHH138 pKa = 5.73FHH140 pKa = 6.29VLLFVEE146 pKa = 5.23KK147 pKa = 10.63LPKK150 pKa = 9.97DD151 pKa = 4.02DD152 pKa = 5.36YY153 pKa = 12.05NDD155 pKa = 3.49ILNLEE160 pKa = 4.77SYY162 pKa = 10.62LSSLFLRR169 pKa = 11.84RR170 pKa = 11.84WQRR173 pKa = 11.84NYY175 pKa = 10.93GSTRR179 pKa = 11.84NPDD182 pKa = 3.31YY183 pKa = 11.28RR184 pKa = 11.84NLCTFVDD191 pKa = 3.58KK192 pKa = 10.75STLFRR197 pKa = 11.84KK198 pKa = 7.64QTNFDD203 pKa = 3.33LHH205 pKa = 5.76YY206 pKa = 10.86VKK208 pKa = 10.54PDD210 pKa = 3.32VNNGISSVVYY220 pKa = 9.58YY221 pKa = 10.42CMKK224 pKa = 10.85YY225 pKa = 8.33MLKK228 pKa = 10.37EE229 pKa = 3.99SSHH232 pKa = 6.7DD233 pKa = 3.99SKK235 pKa = 11.43LHH237 pKa = 5.26YY238 pKa = 10.03ALKK241 pKa = 10.67SSLPPGEE248 pKa = 4.94AAQSWSVVRR257 pKa = 11.84SKK259 pKa = 10.62RR260 pKa = 11.84ACSLDD265 pKa = 3.39FGRR268 pKa = 11.84KK269 pKa = 8.89SDD271 pKa = 3.65PSVLKK276 pKa = 10.51RR277 pKa = 11.84IHH279 pKa = 7.54DD280 pKa = 4.31DD281 pKa = 3.25VDD283 pKa = 3.04KK284 pKa = 11.21CRR286 pKa = 11.84YY287 pKa = 9.46LYY289 pKa = 7.42NTPVFCYY296 pKa = 9.59PDD298 pKa = 3.79CALTAPLSRR307 pKa = 11.84YY308 pKa = 10.11YY309 pKa = 11.07YY310 pKa = 10.46DD311 pKa = 5.26ALSLDD316 pKa = 3.32QVQYY320 pKa = 11.7YY321 pKa = 10.42NDD323 pKa = 4.97LYY325 pKa = 11.27QSLPPSSSLDD335 pKa = 3.38YY336 pKa = 10.89DD337 pKa = 3.61SRR339 pKa = 11.84LLKK342 pKa = 10.92DD343 pKa = 2.9SDD345 pKa = 3.17KK346 pKa = 10.71FRR348 pKa = 11.84RR349 pKa = 11.84LKK351 pKa = 10.16TDD353 pKa = 3.13VFSEE357 pKa = 4.06NVNIYY362 pKa = 10.89EE363 pKa = 4.68NILL366 pKa = 3.49
Molecular weight: 42.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1745 |
73 |
654 |
290.8 |
32.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.986 ± 1.052 | 1.605 ± 0.463 |
7.679 ± 0.365 | 3.381 ± 0.469 |
6.991 ± 0.909 | 4.814 ± 0.854 |
2.006 ± 0.416 | 4.928 ± 0.571 |
4.527 ± 0.677 | 10.372 ± 1.317 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.777 ± 0.214 | 5.272 ± 0.601 |
4.069 ± 0.572 | 3.61 ± 0.969 |
5.1 ± 0.429 | 13.123 ± 0.749 |
3.725 ± 0.409 | 5.903 ± 0.889 |
0.573 ± 0.151 | 5.559 ± 0.773 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
