
Isoptericola sp. CG 20/1183
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Promicromonosporaceae; Isoptericola; unclassified Isoptericola
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
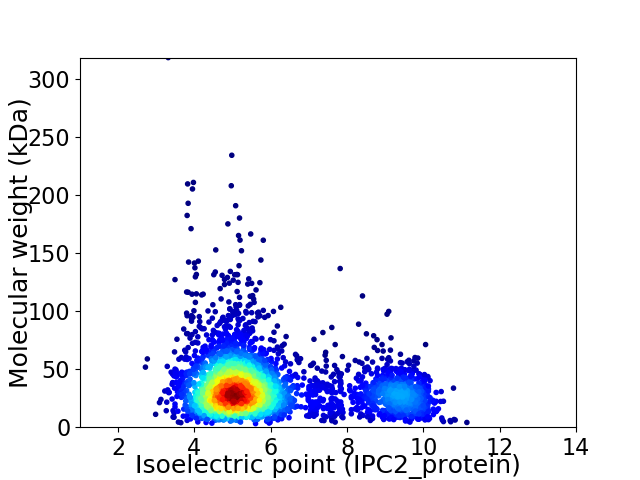
Virtual 2D-PAGE plot for 3485 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T0Y435|A0A2T0Y435_9MICO Xanthine dehydrogenase accessory factor OS=Isoptericola sp. CG 20/1183 OX=1881052 GN=BCE75_104201 PE=4 SV=1
MM1 pKa = 8.1RR2 pKa = 11.84SRR4 pKa = 11.84SLRR7 pKa = 11.84PRR9 pKa = 11.84DD10 pKa = 3.48EE11 pKa = 4.23AVSRR15 pKa = 11.84PVAVAATLALAAVPLLGATTTASAAEE41 pKa = 4.27RR42 pKa = 11.84VDD44 pKa = 3.59NPYY47 pKa = 10.81VGADD51 pKa = 3.39QYY53 pKa = 11.91VNSYY57 pKa = 8.45WSEE60 pKa = 4.05NVVDD64 pKa = 4.78AAAEE68 pKa = 4.08AGGDD72 pKa = 3.87LGQQMLTLADD82 pKa = 3.94TPTSVWMDD90 pKa = 3.21RR91 pKa = 11.84TSAIEE96 pKa = 4.06GNVDD100 pKa = 3.6GPGLRR105 pKa = 11.84YY106 pKa = 9.84HH107 pKa = 7.37LDD109 pKa = 3.34AALEE113 pKa = 4.08QQQGDD118 pKa = 3.64TPMVFNLVIYY128 pKa = 8.83NLPGRR133 pKa = 11.84DD134 pKa = 3.75CYY136 pKa = 11.63ALASNGLLDD145 pKa = 3.93PTPEE149 pKa = 4.13GMAEE153 pKa = 4.08YY154 pKa = 9.99KK155 pKa = 10.52NEE157 pKa = 4.42YY158 pKa = 9.3IDD160 pKa = 5.26VIADD164 pKa = 3.51MLAEE168 pKa = 4.11PQYY171 pKa = 11.69ADD173 pKa = 3.58LRR175 pKa = 11.84VVATIEE181 pKa = 4.21PDD183 pKa = 3.36SLPNLVTNMSEE194 pKa = 4.43PKK196 pKa = 9.45CQEE199 pKa = 3.48SAPYY203 pKa = 9.47YY204 pKa = 10.45RR205 pKa = 11.84EE206 pKa = 3.89GTAYY210 pKa = 10.45ALDD213 pKa = 3.72RR214 pKa = 11.84LYY216 pKa = 11.08EE217 pKa = 4.26AGNVYY222 pKa = 10.54SYY224 pKa = 11.27IDD226 pKa = 3.57AAHH229 pKa = 6.66AGWLGWDD236 pKa = 3.86SNAGPAVDD244 pKa = 3.87VFQEE248 pKa = 4.49VVTSTEE254 pKa = 3.54HH255 pKa = 7.32GYY257 pKa = 8.65DD258 pKa = 3.3TVAGFVTNTANSTPLHH274 pKa = 6.05EE275 pKa = 5.43PFLEE279 pKa = 4.42DD280 pKa = 3.75EE281 pKa = 4.74PFGSGGGTQVRR292 pKa = 11.84QADD295 pKa = 5.09FYY297 pKa = 11.35QWNSDD302 pKa = 3.31FDD304 pKa = 3.95EE305 pKa = 5.15LDD307 pKa = 3.4WTEE310 pKa = 4.25HH311 pKa = 6.16LYY313 pKa = 11.21SLAVAAGFPSDD324 pKa = 3.31IGMLIDD330 pKa = 3.66TSRR333 pKa = 11.84NGWGGPDD340 pKa = 3.21RR341 pKa = 11.84PTAEE345 pKa = 4.48SSSSTLNTYY354 pKa = 9.97VDD356 pKa = 3.67EE357 pKa = 4.55SRR359 pKa = 11.84VDD361 pKa = 3.34RR362 pKa = 11.84RR363 pKa = 11.84THH365 pKa = 6.28RR366 pKa = 11.84GAWCNPAGAGLGEE379 pKa = 4.53RR380 pKa = 11.84PTVNPTDD387 pKa = 3.8NPASHH392 pKa = 7.3LDD394 pKa = 3.17AYY396 pKa = 10.95VWVKK400 pKa = 10.59PPGEE404 pKa = 3.89SDD406 pKa = 3.57GNSEE410 pKa = 5.58EE411 pKa = 4.44IPNDD415 pKa = 3.09QGKK418 pKa = 9.72SFDD421 pKa = 4.04RR422 pKa = 11.84MCDD425 pKa = 3.25PTFASPKK432 pKa = 10.1LDD434 pKa = 3.43GKK436 pKa = 9.43LTGALGGAPLAGQWFQDD453 pKa = 3.52QFEE456 pKa = 4.42MLVTNAYY463 pKa = 9.28PPIDD467 pKa = 4.56GGSVDD472 pKa = 5.2PDD474 pKa = 3.82TEE476 pKa = 4.66APTAPADD483 pKa = 3.7LAATGTTSTSTDD495 pKa = 3.78LTWGASSDD503 pKa = 3.76DD504 pKa = 3.84TGVTRR509 pKa = 11.84YY510 pKa = 7.66TVLVDD515 pKa = 3.36GAEE518 pKa = 4.4AGTSATTSYY527 pKa = 10.03TLTGLTPSTTYY538 pKa = 10.58EE539 pKa = 4.08VTVTARR545 pKa = 11.84DD546 pKa = 3.28AAGNVSAASAPVTVTTKK563 pKa = 10.32EE564 pKa = 3.92ATGGEE569 pKa = 4.35DD570 pKa = 3.21TQAPTVPTGLAAGEE584 pKa = 4.39VTSSSVEE591 pKa = 4.18LTWAAADD598 pKa = 4.54DD599 pKa = 4.06DD600 pKa = 4.57TGVAGYY606 pKa = 9.65RR607 pKa = 11.84VHH609 pKa = 7.85RR610 pKa = 11.84DD611 pKa = 2.74GDD613 pKa = 3.88LVADD617 pKa = 3.91VTGTSATDD625 pKa = 2.96SGLAADD631 pKa = 4.02TAYY634 pKa = 10.83SYY636 pKa = 10.75TVTAYY641 pKa = 10.27DD642 pKa = 3.37AAGNASAASDD652 pKa = 3.72ALSVTTEE659 pKa = 4.04EE660 pKa = 5.34GGTTPTGGCTVDD672 pKa = 3.41YY673 pKa = 10.81SANSWNTGFTGSFVVTNDD691 pKa = 2.42SDD693 pKa = 3.85AAIDD697 pKa = 3.68GWEE700 pKa = 4.11LTFTFADD707 pKa = 4.29GQTVDD712 pKa = 4.58QIWSAQASQSGSAVTITPAAWSTTIPAGGSVTFGFNGSHH751 pKa = 6.53SGTNTAPDD759 pKa = 4.07DD760 pKa = 3.8FALDD764 pKa = 4.06GVACDD769 pKa = 3.63IAA771 pKa = 5.89
MM1 pKa = 8.1RR2 pKa = 11.84SRR4 pKa = 11.84SLRR7 pKa = 11.84PRR9 pKa = 11.84DD10 pKa = 3.48EE11 pKa = 4.23AVSRR15 pKa = 11.84PVAVAATLALAAVPLLGATTTASAAEE41 pKa = 4.27RR42 pKa = 11.84VDD44 pKa = 3.59NPYY47 pKa = 10.81VGADD51 pKa = 3.39QYY53 pKa = 11.91VNSYY57 pKa = 8.45WSEE60 pKa = 4.05NVVDD64 pKa = 4.78AAAEE68 pKa = 4.08AGGDD72 pKa = 3.87LGQQMLTLADD82 pKa = 3.94TPTSVWMDD90 pKa = 3.21RR91 pKa = 11.84TSAIEE96 pKa = 4.06GNVDD100 pKa = 3.6GPGLRR105 pKa = 11.84YY106 pKa = 9.84HH107 pKa = 7.37LDD109 pKa = 3.34AALEE113 pKa = 4.08QQQGDD118 pKa = 3.64TPMVFNLVIYY128 pKa = 8.83NLPGRR133 pKa = 11.84DD134 pKa = 3.75CYY136 pKa = 11.63ALASNGLLDD145 pKa = 3.93PTPEE149 pKa = 4.13GMAEE153 pKa = 4.08YY154 pKa = 9.99KK155 pKa = 10.52NEE157 pKa = 4.42YY158 pKa = 9.3IDD160 pKa = 5.26VIADD164 pKa = 3.51MLAEE168 pKa = 4.11PQYY171 pKa = 11.69ADD173 pKa = 3.58LRR175 pKa = 11.84VVATIEE181 pKa = 4.21PDD183 pKa = 3.36SLPNLVTNMSEE194 pKa = 4.43PKK196 pKa = 9.45CQEE199 pKa = 3.48SAPYY203 pKa = 9.47YY204 pKa = 10.45RR205 pKa = 11.84EE206 pKa = 3.89GTAYY210 pKa = 10.45ALDD213 pKa = 3.72RR214 pKa = 11.84LYY216 pKa = 11.08EE217 pKa = 4.26AGNVYY222 pKa = 10.54SYY224 pKa = 11.27IDD226 pKa = 3.57AAHH229 pKa = 6.66AGWLGWDD236 pKa = 3.86SNAGPAVDD244 pKa = 3.87VFQEE248 pKa = 4.49VVTSTEE254 pKa = 3.54HH255 pKa = 7.32GYY257 pKa = 8.65DD258 pKa = 3.3TVAGFVTNTANSTPLHH274 pKa = 6.05EE275 pKa = 5.43PFLEE279 pKa = 4.42DD280 pKa = 3.75EE281 pKa = 4.74PFGSGGGTQVRR292 pKa = 11.84QADD295 pKa = 5.09FYY297 pKa = 11.35QWNSDD302 pKa = 3.31FDD304 pKa = 3.95EE305 pKa = 5.15LDD307 pKa = 3.4WTEE310 pKa = 4.25HH311 pKa = 6.16LYY313 pKa = 11.21SLAVAAGFPSDD324 pKa = 3.31IGMLIDD330 pKa = 3.66TSRR333 pKa = 11.84NGWGGPDD340 pKa = 3.21RR341 pKa = 11.84PTAEE345 pKa = 4.48SSSSTLNTYY354 pKa = 9.97VDD356 pKa = 3.67EE357 pKa = 4.55SRR359 pKa = 11.84VDD361 pKa = 3.34RR362 pKa = 11.84RR363 pKa = 11.84THH365 pKa = 6.28RR366 pKa = 11.84GAWCNPAGAGLGEE379 pKa = 4.53RR380 pKa = 11.84PTVNPTDD387 pKa = 3.8NPASHH392 pKa = 7.3LDD394 pKa = 3.17AYY396 pKa = 10.95VWVKK400 pKa = 10.59PPGEE404 pKa = 3.89SDD406 pKa = 3.57GNSEE410 pKa = 5.58EE411 pKa = 4.44IPNDD415 pKa = 3.09QGKK418 pKa = 9.72SFDD421 pKa = 4.04RR422 pKa = 11.84MCDD425 pKa = 3.25PTFASPKK432 pKa = 10.1LDD434 pKa = 3.43GKK436 pKa = 9.43LTGALGGAPLAGQWFQDD453 pKa = 3.52QFEE456 pKa = 4.42MLVTNAYY463 pKa = 9.28PPIDD467 pKa = 4.56GGSVDD472 pKa = 5.2PDD474 pKa = 3.82TEE476 pKa = 4.66APTAPADD483 pKa = 3.7LAATGTTSTSTDD495 pKa = 3.78LTWGASSDD503 pKa = 3.76DD504 pKa = 3.84TGVTRR509 pKa = 11.84YY510 pKa = 7.66TVLVDD515 pKa = 3.36GAEE518 pKa = 4.4AGTSATTSYY527 pKa = 10.03TLTGLTPSTTYY538 pKa = 10.58EE539 pKa = 4.08VTVTARR545 pKa = 11.84DD546 pKa = 3.28AAGNVSAASAPVTVTTKK563 pKa = 10.32EE564 pKa = 3.92ATGGEE569 pKa = 4.35DD570 pKa = 3.21TQAPTVPTGLAAGEE584 pKa = 4.39VTSSSVEE591 pKa = 4.18LTWAAADD598 pKa = 4.54DD599 pKa = 4.06DD600 pKa = 4.57TGVAGYY606 pKa = 9.65RR607 pKa = 11.84VHH609 pKa = 7.85RR610 pKa = 11.84DD611 pKa = 2.74GDD613 pKa = 3.88LVADD617 pKa = 3.91VTGTSATDD625 pKa = 2.96SGLAADD631 pKa = 4.02TAYY634 pKa = 10.83SYY636 pKa = 10.75TVTAYY641 pKa = 10.27DD642 pKa = 3.37AAGNASAASDD652 pKa = 3.72ALSVTTEE659 pKa = 4.04EE660 pKa = 5.34GGTTPTGGCTVDD672 pKa = 3.41YY673 pKa = 10.81SANSWNTGFTGSFVVTNDD691 pKa = 2.42SDD693 pKa = 3.85AAIDD697 pKa = 3.68GWEE700 pKa = 4.11LTFTFADD707 pKa = 4.29GQTVDD712 pKa = 4.58QIWSAQASQSGSAVTITPAAWSTTIPAGGSVTFGFNGSHH751 pKa = 6.53SGTNTAPDD759 pKa = 4.07DD760 pKa = 3.8FALDD764 pKa = 4.06GVACDD769 pKa = 3.63IAA771 pKa = 5.89
Molecular weight: 80.51 kDa
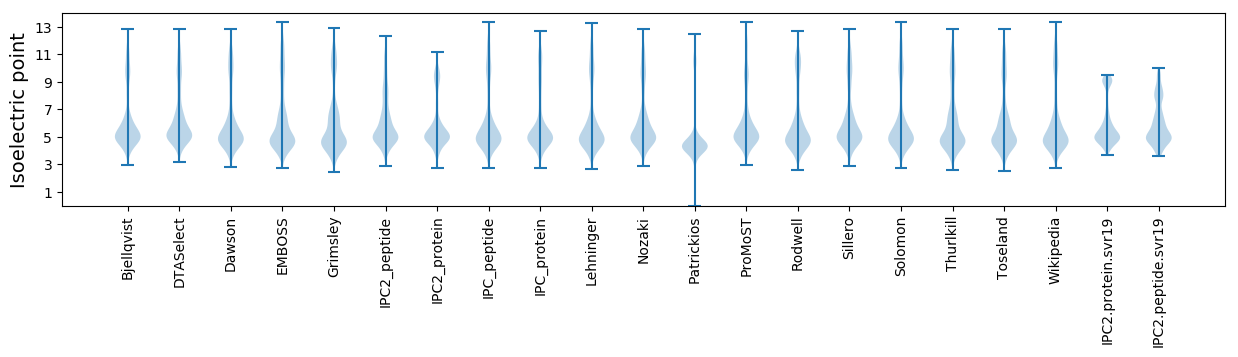
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T0Y6M9|A0A2T0Y6M9_9MICO Isochorismate synthase OS=Isoptericola sp. CG 20/1183 OX=1881052 GN=BCE75_102398 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1184991 |
30 |
3095 |
340.0 |
36.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.14 ± 0.063 | 0.544 ± 0.01 |
6.776 ± 0.043 | 5.703 ± 0.037 |
2.555 ± 0.024 | 9.474 ± 0.038 |
2.166 ± 0.02 | 2.797 ± 0.026 |
1.402 ± 0.027 | 10.068 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.611 ± 0.015 | 1.482 ± 0.022 |
5.889 ± 0.035 | 2.604 ± 0.021 |
7.842 ± 0.053 | 4.99 ± 0.03 |
6.366 ± 0.033 | 10.15 ± 0.043 |
1.59 ± 0.019 | 1.851 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
