
Bandicoot papillomatosis carcinomatosis virus type 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 5.33
Get precalculated fractions of proteins
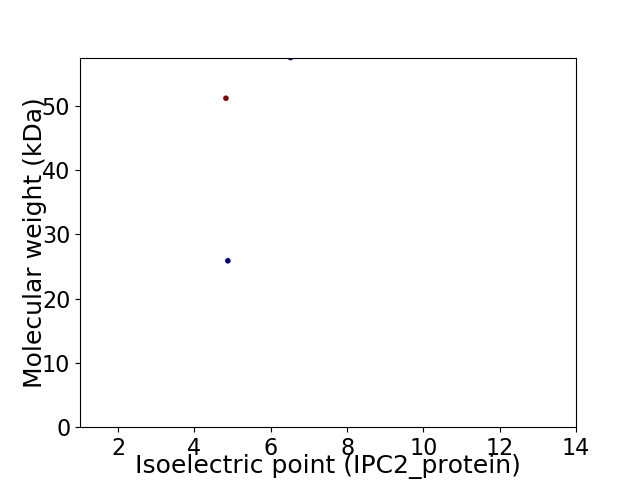
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B3FN93|B3FN93_9PAPI Small t antigen OS=Bandicoot papillomatosis carcinomatosis virus type 2 OX=500654 GN=stag PE=4 SV=1
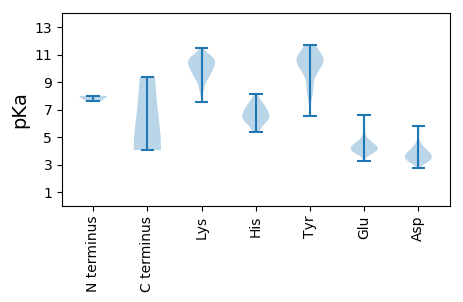
MM1 pKa = 7.99DD2 pKa = 4.52NSDD5 pKa = 3.6YY6 pKa = 11.67LEE8 pKa = 3.98FLEE11 pKa = 6.58LIGCDD16 pKa = 3.34TYY18 pKa = 11.57DD19 pKa = 3.59GPEE22 pKa = 4.2NIKK25 pKa = 10.5KK26 pKa = 10.21KK27 pKa = 9.46YY28 pKa = 9.95RR29 pKa = 11.84KK30 pKa = 9.23AALKK34 pKa = 9.58NHH36 pKa = 6.73PDD38 pKa = 3.31KK39 pKa = 11.47GGSANKK45 pKa = 9.24MKK47 pKa = 10.64RR48 pKa = 11.84LNEE51 pKa = 3.74LNQIYY56 pKa = 7.84TEE58 pKa = 4.62CGPPQNKK65 pKa = 9.26RR66 pKa = 11.84YY67 pKa = 9.58RR68 pKa = 11.84PSSHH72 pKa = 7.04YY73 pKa = 10.47EE74 pKa = 3.83EE75 pKa = 5.59EE76 pKa = 5.04DD77 pKa = 3.55LFCYY81 pKa = 10.43EE82 pKa = 4.92SGGEE86 pKa = 4.49DD87 pKa = 3.77EE88 pKa = 6.13PDD90 pKa = 3.14TCSYY94 pKa = 10.69NRR96 pKa = 11.84SDD98 pKa = 3.87SGFSEE103 pKa = 4.59GNYY106 pKa = 6.55TTPDD110 pKa = 3.38PSGFSASNVSFSQSCRR126 pKa = 11.84NVKK129 pKa = 10.2LLLDD133 pKa = 3.36ITEE136 pKa = 4.37TLGHH140 pKa = 6.31GRR142 pKa = 11.84HH143 pKa = 6.12CAHH146 pKa = 6.61TLEE149 pKa = 6.3AIDD152 pKa = 5.28KK153 pKa = 8.49NNKK156 pKa = 8.35MDD158 pKa = 3.49WEE160 pKa = 4.35LFDD163 pKa = 4.47TFCATPGNHH172 pKa = 7.08HH173 pKa = 6.31LTIMFIEE180 pKa = 4.34HH181 pKa = 7.39CYY183 pKa = 9.98MKK185 pKa = 10.81LKK187 pKa = 10.35DD188 pKa = 3.95WQWSLMNFVNYY199 pKa = 9.74GPSYY203 pKa = 10.75LEE205 pKa = 4.36EE206 pKa = 4.48IKK208 pKa = 10.74EE209 pKa = 3.89AANAINWSQLDD220 pKa = 3.67DD221 pKa = 4.18MIFSS225 pKa = 4.14
MM1 pKa = 7.99DD2 pKa = 4.52NSDD5 pKa = 3.6YY6 pKa = 11.67LEE8 pKa = 3.98FLEE11 pKa = 6.58LIGCDD16 pKa = 3.34TYY18 pKa = 11.57DD19 pKa = 3.59GPEE22 pKa = 4.2NIKK25 pKa = 10.5KK26 pKa = 10.21KK27 pKa = 9.46YY28 pKa = 9.95RR29 pKa = 11.84KK30 pKa = 9.23AALKK34 pKa = 9.58NHH36 pKa = 6.73PDD38 pKa = 3.31KK39 pKa = 11.47GGSANKK45 pKa = 9.24MKK47 pKa = 10.64RR48 pKa = 11.84LNEE51 pKa = 3.74LNQIYY56 pKa = 7.84TEE58 pKa = 4.62CGPPQNKK65 pKa = 9.26RR66 pKa = 11.84YY67 pKa = 9.58RR68 pKa = 11.84PSSHH72 pKa = 7.04YY73 pKa = 10.47EE74 pKa = 3.83EE75 pKa = 5.59EE76 pKa = 5.04DD77 pKa = 3.55LFCYY81 pKa = 10.43EE82 pKa = 4.92SGGEE86 pKa = 4.49DD87 pKa = 3.77EE88 pKa = 6.13PDD90 pKa = 3.14TCSYY94 pKa = 10.69NRR96 pKa = 11.84SDD98 pKa = 3.87SGFSEE103 pKa = 4.59GNYY106 pKa = 6.55TTPDD110 pKa = 3.38PSGFSASNVSFSQSCRR126 pKa = 11.84NVKK129 pKa = 10.2LLLDD133 pKa = 3.36ITEE136 pKa = 4.37TLGHH140 pKa = 6.31GRR142 pKa = 11.84HH143 pKa = 6.12CAHH146 pKa = 6.61TLEE149 pKa = 6.3AIDD152 pKa = 5.28KK153 pKa = 8.49NNKK156 pKa = 8.35MDD158 pKa = 3.49WEE160 pKa = 4.35LFDD163 pKa = 4.47TFCATPGNHH172 pKa = 7.08HH173 pKa = 6.31LTIMFIEE180 pKa = 4.34HH181 pKa = 7.39CYY183 pKa = 9.98MKK185 pKa = 10.81LKK187 pKa = 10.35DD188 pKa = 3.95WQWSLMNFVNYY199 pKa = 9.74GPSYY203 pKa = 10.75LEE205 pKa = 4.36EE206 pKa = 4.48IKK208 pKa = 10.74EE209 pKa = 3.89AANAINWSQLDD220 pKa = 3.67DD221 pKa = 4.18MIFSS225 pKa = 4.14
Molecular weight: 25.87 kDa
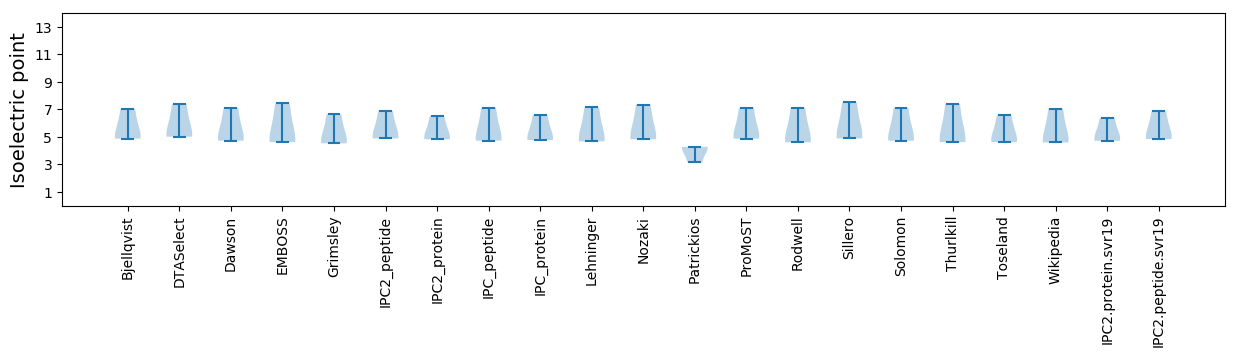
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B3FN93|B3FN93_9PAPI Small t antigen OS=Bandicoot papillomatosis carcinomatosis virus type 2 OX=500654 GN=stag PE=4 SV=1
MM1 pKa = 7.93AIWQPNGKK9 pKa = 9.93VYY11 pKa = 10.68LPPSAPVARR20 pKa = 11.84ILNTEE25 pKa = 4.21EE26 pKa = 3.93YY27 pKa = 9.77IQRR30 pKa = 11.84TNLFYY35 pKa = 10.66HH36 pKa = 6.99ANTEE40 pKa = 3.95RR41 pKa = 11.84LLTVGHH47 pKa = 7.43PYY49 pKa = 10.94YY50 pKa = 10.38EE51 pKa = 4.34IKK53 pKa = 10.89DD54 pKa = 3.69NLDD57 pKa = 3.3PSKK60 pKa = 10.77IGVPKK65 pKa = 10.6VSANQYY71 pKa = 8.75RR72 pKa = 11.84VFRR75 pKa = 11.84IKK77 pKa = 10.92LPDD80 pKa = 4.01PNRR83 pKa = 11.84FALGDD88 pKa = 3.44KK89 pKa = 10.45SLYY92 pKa = 10.63DD93 pKa = 3.64PEE95 pKa = 4.66KK96 pKa = 10.7EE97 pKa = 3.83RR98 pKa = 11.84LVWGVRR104 pKa = 11.84GLEE107 pKa = 4.05IEE109 pKa = 4.3RR110 pKa = 11.84GQPLNVGSSGNVLYY124 pKa = 11.19NKK126 pKa = 10.15FKK128 pKa = 10.46DD129 pKa = 3.7TEE131 pKa = 4.25NPNNYY136 pKa = 9.68AKK138 pKa = 10.57EE139 pKa = 4.47GEE141 pKa = 4.28DD142 pKa = 3.25EE143 pKa = 4.24RR144 pKa = 11.84QNVSHH149 pKa = 7.51DD150 pKa = 3.78PKK152 pKa = 10.69QMQMLLVGCKK162 pKa = 9.37PATGEE167 pKa = 3.82HH168 pKa = 6.02WDD170 pKa = 3.59AAKK173 pKa = 9.44TCAEE177 pKa = 4.09RR178 pKa = 11.84PPNKK182 pKa = 9.82GDD184 pKa = 3.8CPPLEE189 pKa = 4.32LVNSIIEE196 pKa = 4.65DD197 pKa = 3.37GDD199 pKa = 4.03MIDD202 pKa = 4.24IGYY205 pKa = 10.86GNLNFKK211 pKa = 7.55TLCGNKK217 pKa = 9.67SDD219 pKa = 4.25VPLDD223 pKa = 3.35INTTTTKK230 pKa = 10.77YY231 pKa = 10.55PDD233 pKa = 3.9FLKK236 pKa = 8.54MTSDD240 pKa = 3.44QYY242 pKa = 11.69GDD244 pKa = 3.6SLFFYY249 pKa = 9.09TKK251 pKa = 10.34RR252 pKa = 11.84EE253 pKa = 3.93STYY256 pKa = 11.15ARR258 pKa = 11.84HH259 pKa = 5.5FWTRR263 pKa = 11.84GGTPGDD269 pKa = 4.73PIPEE273 pKa = 4.0EE274 pKa = 4.64LFVNPSDD281 pKa = 3.66NQKK284 pKa = 10.0ILGSSTYY291 pKa = 8.56FTVPSGSLVSSEE303 pKa = 3.88SQLFNRR309 pKa = 11.84PYY311 pKa = 9.9WLQKK315 pKa = 10.1ATGQNNGICWNNDD328 pKa = 2.76LFITVVDD335 pKa = 3.59NTRR338 pKa = 11.84NCNFTISVFNKK349 pKa = 9.54ADD351 pKa = 3.35SSQGQTYY358 pKa = 10.26KK359 pKa = 10.75SSNYY363 pKa = 10.51NMFLRR368 pKa = 11.84HH369 pKa = 5.36VEE371 pKa = 3.94EE372 pKa = 4.95FEE374 pKa = 4.85LNFIFEE380 pKa = 4.24ICKK383 pKa = 10.15VPLKK387 pKa = 10.77ADD389 pKa = 3.45VLAHH393 pKa = 6.42INAMDD398 pKa = 3.85PRR400 pKa = 11.84ILEE403 pKa = 4.24DD404 pKa = 3.02WSLGFVPSNNFPLEE418 pKa = 4.04DD419 pKa = 3.31KK420 pKa = 11.15YY421 pKa = 11.57RR422 pKa = 11.84FINSLATRR430 pKa = 11.84CPDD433 pKa = 3.48KK434 pKa = 10.29EE435 pKa = 4.59TPKK438 pKa = 10.66EE439 pKa = 3.94KK440 pKa = 10.11EE441 pKa = 3.59DD442 pKa = 3.89PYY444 pKa = 10.97RR445 pKa = 11.84SNTFWTVEE453 pKa = 3.82LEE455 pKa = 3.97EE456 pKa = 4.57RR457 pKa = 11.84MSSEE461 pKa = 3.91LDD463 pKa = 3.2QYY465 pKa = 11.36PLGRR469 pKa = 11.84KK470 pKa = 8.67FLFQSNISSINTVAKK485 pKa = 9.24PRR487 pKa = 11.84KK488 pKa = 9.31RR489 pKa = 11.84KK490 pKa = 8.58ATTPTTKK497 pKa = 9.76TKK499 pKa = 10.2RR500 pKa = 11.84RR501 pKa = 11.84KK502 pKa = 9.36
MM1 pKa = 7.93AIWQPNGKK9 pKa = 9.93VYY11 pKa = 10.68LPPSAPVARR20 pKa = 11.84ILNTEE25 pKa = 4.21EE26 pKa = 3.93YY27 pKa = 9.77IQRR30 pKa = 11.84TNLFYY35 pKa = 10.66HH36 pKa = 6.99ANTEE40 pKa = 3.95RR41 pKa = 11.84LLTVGHH47 pKa = 7.43PYY49 pKa = 10.94YY50 pKa = 10.38EE51 pKa = 4.34IKK53 pKa = 10.89DD54 pKa = 3.69NLDD57 pKa = 3.3PSKK60 pKa = 10.77IGVPKK65 pKa = 10.6VSANQYY71 pKa = 8.75RR72 pKa = 11.84VFRR75 pKa = 11.84IKK77 pKa = 10.92LPDD80 pKa = 4.01PNRR83 pKa = 11.84FALGDD88 pKa = 3.44KK89 pKa = 10.45SLYY92 pKa = 10.63DD93 pKa = 3.64PEE95 pKa = 4.66KK96 pKa = 10.7EE97 pKa = 3.83RR98 pKa = 11.84LVWGVRR104 pKa = 11.84GLEE107 pKa = 4.05IEE109 pKa = 4.3RR110 pKa = 11.84GQPLNVGSSGNVLYY124 pKa = 11.19NKK126 pKa = 10.15FKK128 pKa = 10.46DD129 pKa = 3.7TEE131 pKa = 4.25NPNNYY136 pKa = 9.68AKK138 pKa = 10.57EE139 pKa = 4.47GEE141 pKa = 4.28DD142 pKa = 3.25EE143 pKa = 4.24RR144 pKa = 11.84QNVSHH149 pKa = 7.51DD150 pKa = 3.78PKK152 pKa = 10.69QMQMLLVGCKK162 pKa = 9.37PATGEE167 pKa = 3.82HH168 pKa = 6.02WDD170 pKa = 3.59AAKK173 pKa = 9.44TCAEE177 pKa = 4.09RR178 pKa = 11.84PPNKK182 pKa = 9.82GDD184 pKa = 3.8CPPLEE189 pKa = 4.32LVNSIIEE196 pKa = 4.65DD197 pKa = 3.37GDD199 pKa = 4.03MIDD202 pKa = 4.24IGYY205 pKa = 10.86GNLNFKK211 pKa = 7.55TLCGNKK217 pKa = 9.67SDD219 pKa = 4.25VPLDD223 pKa = 3.35INTTTTKK230 pKa = 10.77YY231 pKa = 10.55PDD233 pKa = 3.9FLKK236 pKa = 8.54MTSDD240 pKa = 3.44QYY242 pKa = 11.69GDD244 pKa = 3.6SLFFYY249 pKa = 9.09TKK251 pKa = 10.34RR252 pKa = 11.84EE253 pKa = 3.93STYY256 pKa = 11.15ARR258 pKa = 11.84HH259 pKa = 5.5FWTRR263 pKa = 11.84GGTPGDD269 pKa = 4.73PIPEE273 pKa = 4.0EE274 pKa = 4.64LFVNPSDD281 pKa = 3.66NQKK284 pKa = 10.0ILGSSTYY291 pKa = 8.56FTVPSGSLVSSEE303 pKa = 3.88SQLFNRR309 pKa = 11.84PYY311 pKa = 9.9WLQKK315 pKa = 10.1ATGQNNGICWNNDD328 pKa = 2.76LFITVVDD335 pKa = 3.59NTRR338 pKa = 11.84NCNFTISVFNKK349 pKa = 9.54ADD351 pKa = 3.35SSQGQTYY358 pKa = 10.26KK359 pKa = 10.75SSNYY363 pKa = 10.51NMFLRR368 pKa = 11.84HH369 pKa = 5.36VEE371 pKa = 3.94EE372 pKa = 4.95FEE374 pKa = 4.85LNFIFEE380 pKa = 4.24ICKK383 pKa = 10.15VPLKK387 pKa = 10.77ADD389 pKa = 3.45VLAHH393 pKa = 6.42INAMDD398 pKa = 3.85PRR400 pKa = 11.84ILEE403 pKa = 4.24DD404 pKa = 3.02WSLGFVPSNNFPLEE418 pKa = 4.04DD419 pKa = 3.31KK420 pKa = 11.15YY421 pKa = 11.57RR422 pKa = 11.84FINSLATRR430 pKa = 11.84CPDD433 pKa = 3.48KK434 pKa = 10.29EE435 pKa = 4.59TPKK438 pKa = 10.66EE439 pKa = 3.94KK440 pKa = 10.11EE441 pKa = 3.59DD442 pKa = 3.89PYY444 pKa = 10.97RR445 pKa = 11.84SNTFWTVEE453 pKa = 3.82LEE455 pKa = 3.97EE456 pKa = 4.57RR457 pKa = 11.84MSSEE461 pKa = 3.91LDD463 pKa = 3.2QYY465 pKa = 11.36PLGRR469 pKa = 11.84KK470 pKa = 8.67FLFQSNISSINTVAKK485 pKa = 9.24PRR487 pKa = 11.84KK488 pKa = 9.31RR489 pKa = 11.84KK490 pKa = 8.58ATTPTTKK497 pKa = 9.76TKK499 pKa = 10.2RR500 pKa = 11.84RR501 pKa = 11.84KK502 pKa = 9.36
Molecular weight: 57.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1196 |
225 |
502 |
398.7 |
44.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.264 ± 0.137 | 1.505 ± 0.583 |
6.689 ± 0.359 | 6.773 ± 0.428 |
4.515 ± 0.24 | 6.773 ± 0.707 |
1.672 ± 0.478 | 6.187 ± 1.033 |
5.769 ± 0.871 | 7.776 ± 0.138 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.756 ± 0.347 | 6.856 ± 0.869 |
7.191 ± 0.758 | 2.926 ± 0.188 |
5.1 ± 0.506 | 7.609 ± 0.412 |
6.438 ± 0.391 | 5.435 ± 1.185 |
1.003 ± 0.442 | 3.763 ± 0.558 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
