
Lepus americanus faeces associated genomovirus SHP9
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus lepam3
Average proteome isoelectric point is 7.33
Get precalculated fractions of proteins
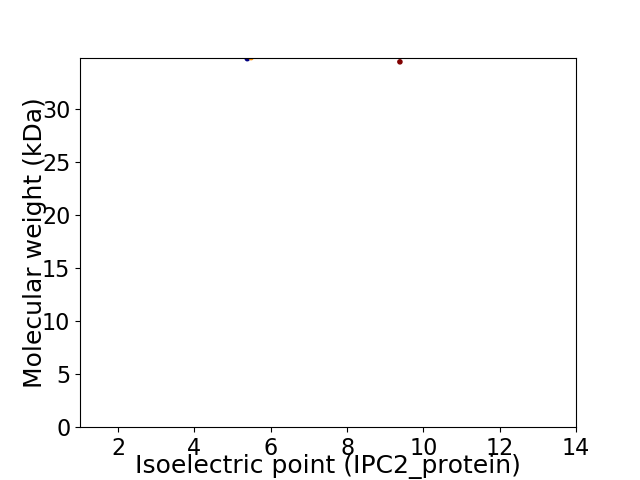
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CJ37|A0A2Z5CJ37_9VIRU Capsid protein OS=Lepus americanus faeces associated genomovirus SHP9 OX=2219121 PE=4 SV=1
MM1 pKa = 7.82PFACNARR8 pKa = 11.84YY9 pKa = 8.97FLVTYY14 pKa = 9.34SHH16 pKa = 6.96VEE18 pKa = 3.77PLDD21 pKa = 3.62PFAVVEE27 pKa = 4.22HH28 pKa = 6.52FGNLGAEE35 pKa = 4.39VIVSLEE41 pKa = 4.23SYY43 pKa = 8.12NTTLGVHH50 pKa = 5.09YY51 pKa = 10.36HH52 pKa = 5.64VFADD56 pKa = 4.64FGRR59 pKa = 11.84KK60 pKa = 8.16FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84TDD67 pKa = 2.68IFDD70 pKa = 3.27VDD72 pKa = 3.91GFHH75 pKa = 7.48PNISPSRR82 pKa = 11.84GTPQAGYY89 pKa = 10.56DD90 pKa = 3.74YY91 pKa = 11.05AVKK94 pKa = 10.52DD95 pKa = 4.0GNVVAGGLARR105 pKa = 11.84PSGVVAAGRR114 pKa = 11.84AAKK117 pKa = 8.68WHH119 pKa = 6.18QILDD123 pKa = 3.47AEE125 pKa = 4.47TRR127 pKa = 11.84DD128 pKa = 3.66EE129 pKa = 5.25FFALCEE135 pKa = 3.97EE136 pKa = 5.06LDD138 pKa = 3.88PEE140 pKa = 4.85RR141 pKa = 11.84LVCSFNQIQKK151 pKa = 9.25FADD154 pKa = 2.73WRR156 pKa = 11.84FAVEE160 pKa = 4.25PKK162 pKa = 10.37PYY164 pKa = 8.97ATPDD168 pKa = 3.43GVFDD172 pKa = 4.28LANYY176 pKa = 10.13GDD178 pKa = 4.02LSEE181 pKa = 4.54SKK183 pKa = 10.8SLILWGPSRR192 pKa = 11.84MGKK195 pKa = 6.86TVWARR200 pKa = 11.84SLGRR204 pKa = 11.84HH205 pKa = 6.15LYY207 pKa = 10.37FGGIFSARR215 pKa = 11.84NLGDD219 pKa = 3.46GGVDD223 pKa = 3.17YY224 pKa = 11.36AVFDD228 pKa = 5.51DD229 pKa = 3.63IAGGIKK235 pKa = 10.03FFPRR239 pKa = 11.84FKK241 pKa = 10.79DD242 pKa = 3.12WLGCQMEE249 pKa = 4.39FMVKK253 pKa = 9.1QMYY256 pKa = 9.85RR257 pKa = 11.84DD258 pKa = 3.38PHH260 pKa = 6.22LFRR263 pKa = 11.84WGKK266 pKa = 7.9PCIWVANTDD275 pKa = 3.43PRR277 pKa = 11.84HH278 pKa = 6.65DD279 pKa = 3.74MTHH282 pKa = 7.65DD283 pKa = 3.17EE284 pKa = 4.56VTWLEE289 pKa = 4.04ANCIFVEE296 pKa = 4.12ISSAIFHH303 pKa = 6.76ANIEE307 pKa = 4.2
MM1 pKa = 7.82PFACNARR8 pKa = 11.84YY9 pKa = 8.97FLVTYY14 pKa = 9.34SHH16 pKa = 6.96VEE18 pKa = 3.77PLDD21 pKa = 3.62PFAVVEE27 pKa = 4.22HH28 pKa = 6.52FGNLGAEE35 pKa = 4.39VIVSLEE41 pKa = 4.23SYY43 pKa = 8.12NTTLGVHH50 pKa = 5.09YY51 pKa = 10.36HH52 pKa = 5.64VFADD56 pKa = 4.64FGRR59 pKa = 11.84KK60 pKa = 8.16FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84TDD67 pKa = 2.68IFDD70 pKa = 3.27VDD72 pKa = 3.91GFHH75 pKa = 7.48PNISPSRR82 pKa = 11.84GTPQAGYY89 pKa = 10.56DD90 pKa = 3.74YY91 pKa = 11.05AVKK94 pKa = 10.52DD95 pKa = 4.0GNVVAGGLARR105 pKa = 11.84PSGVVAAGRR114 pKa = 11.84AAKK117 pKa = 8.68WHH119 pKa = 6.18QILDD123 pKa = 3.47AEE125 pKa = 4.47TRR127 pKa = 11.84DD128 pKa = 3.66EE129 pKa = 5.25FFALCEE135 pKa = 3.97EE136 pKa = 5.06LDD138 pKa = 3.88PEE140 pKa = 4.85RR141 pKa = 11.84LVCSFNQIQKK151 pKa = 9.25FADD154 pKa = 2.73WRR156 pKa = 11.84FAVEE160 pKa = 4.25PKK162 pKa = 10.37PYY164 pKa = 8.97ATPDD168 pKa = 3.43GVFDD172 pKa = 4.28LANYY176 pKa = 10.13GDD178 pKa = 4.02LSEE181 pKa = 4.54SKK183 pKa = 10.8SLILWGPSRR192 pKa = 11.84MGKK195 pKa = 6.86TVWARR200 pKa = 11.84SLGRR204 pKa = 11.84HH205 pKa = 6.15LYY207 pKa = 10.37FGGIFSARR215 pKa = 11.84NLGDD219 pKa = 3.46GGVDD223 pKa = 3.17YY224 pKa = 11.36AVFDD228 pKa = 5.51DD229 pKa = 3.63IAGGIKK235 pKa = 10.03FFPRR239 pKa = 11.84FKK241 pKa = 10.79DD242 pKa = 3.12WLGCQMEE249 pKa = 4.39FMVKK253 pKa = 9.1QMYY256 pKa = 9.85RR257 pKa = 11.84DD258 pKa = 3.38PHH260 pKa = 6.22LFRR263 pKa = 11.84WGKK266 pKa = 7.9PCIWVANTDD275 pKa = 3.43PRR277 pKa = 11.84HH278 pKa = 6.65DD279 pKa = 3.74MTHH282 pKa = 7.65DD283 pKa = 3.17EE284 pKa = 4.56VTWLEE289 pKa = 4.04ANCIFVEE296 pKa = 4.12ISSAIFHH303 pKa = 6.76ANIEE307 pKa = 4.2
Molecular weight: 34.7 kDa
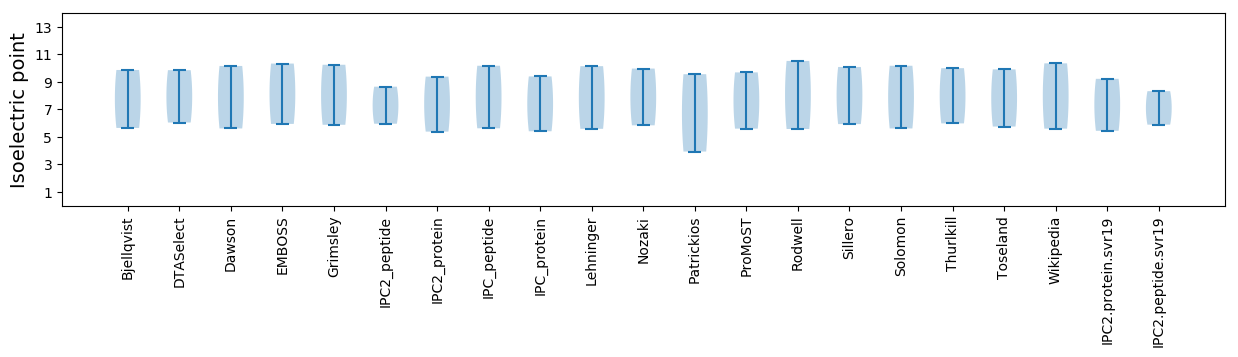
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CJ37|A0A2Z5CJ37_9VIRU Capsid protein OS=Lepus americanus faeces associated genomovirus SHP9 OX=2219121 PE=4 SV=1
MM1 pKa = 7.61AYY3 pKa = 10.1ARR5 pKa = 11.84KK6 pKa = 9.82RR7 pKa = 11.84YY8 pKa = 6.3TRR10 pKa = 11.84KK11 pKa = 9.59KK12 pKa = 7.83GAYY15 pKa = 8.95RR16 pKa = 11.84KK17 pKa = 8.44RR18 pKa = 11.84TATKK22 pKa = 10.06RR23 pKa = 11.84RR24 pKa = 11.84SSYY27 pKa = 10.42AKK29 pKa = 9.43KK30 pKa = 8.94RR31 pKa = 11.84TYY33 pKa = 10.46RR34 pKa = 11.84KK35 pKa = 8.87PAMSKK40 pKa = 10.33KK41 pKa = 10.33RR42 pKa = 11.84ILTVTSTKK50 pKa = 10.43KK51 pKa = 10.25RR52 pKa = 11.84DD53 pKa = 3.45TMLNWTNSTSSTQVGGTTYY72 pKa = 10.97GSAPAVITGGSNAQNCAAFLWCATARR98 pKa = 11.84DD99 pKa = 4.14ATVSSAGGPTTKK111 pKa = 10.42YY112 pKa = 10.89ALAGRR117 pKa = 11.84SATSCYY123 pKa = 9.66MVGLAEE129 pKa = 5.39KK130 pKa = 10.48IEE132 pKa = 4.2IQCNTGMPWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTMKK151 pKa = 10.32GVSLVPAATGSGTQFSDD168 pKa = 4.17FLEE171 pKa = 4.5TSNGYY176 pKa = 8.07MRR178 pKa = 11.84VMNQVNGNPSLDD190 pKa = 3.28PMYY193 pKa = 11.03SLFGALFKK201 pKa = 11.24GQVNSDD207 pKa = 2.95WQDD210 pKa = 2.91PMTAPTDD217 pKa = 3.57NSRR220 pKa = 11.84LSIKK224 pKa = 9.81YY225 pKa = 10.43DD226 pKa = 3.17KK227 pKa = 10.17VTSLASGNEE236 pKa = 3.89DD237 pKa = 3.44GFIRR241 pKa = 11.84SYY243 pKa = 11.1KK244 pKa = 9.57RR245 pKa = 11.84WHH247 pKa = 6.77PMNKK251 pKa = 8.21TLVYY255 pKa = 10.82NDD257 pKa = 4.58DD258 pKa = 3.83EE259 pKa = 5.64LGGGMLSNFKK269 pKa = 10.9SSFGKK274 pKa = 10.48AGMGDD279 pKa = 4.22YY280 pKa = 10.8YY281 pKa = 11.48VVDD284 pKa = 3.75IFRR287 pKa = 11.84ARR289 pKa = 11.84QGSATTDD296 pKa = 3.36QLSVCPEE303 pKa = 3.55ATLYY307 pKa = 9.32WHH309 pKa = 7.08EE310 pKa = 4.26KK311 pKa = 9.3
MM1 pKa = 7.61AYY3 pKa = 10.1ARR5 pKa = 11.84KK6 pKa = 9.82RR7 pKa = 11.84YY8 pKa = 6.3TRR10 pKa = 11.84KK11 pKa = 9.59KK12 pKa = 7.83GAYY15 pKa = 8.95RR16 pKa = 11.84KK17 pKa = 8.44RR18 pKa = 11.84TATKK22 pKa = 10.06RR23 pKa = 11.84RR24 pKa = 11.84SSYY27 pKa = 10.42AKK29 pKa = 9.43KK30 pKa = 8.94RR31 pKa = 11.84TYY33 pKa = 10.46RR34 pKa = 11.84KK35 pKa = 8.87PAMSKK40 pKa = 10.33KK41 pKa = 10.33RR42 pKa = 11.84ILTVTSTKK50 pKa = 10.43KK51 pKa = 10.25RR52 pKa = 11.84DD53 pKa = 3.45TMLNWTNSTSSTQVGGTTYY72 pKa = 10.97GSAPAVITGGSNAQNCAAFLWCATARR98 pKa = 11.84DD99 pKa = 4.14ATVSSAGGPTTKK111 pKa = 10.42YY112 pKa = 10.89ALAGRR117 pKa = 11.84SATSCYY123 pKa = 9.66MVGLAEE129 pKa = 5.39KK130 pKa = 10.48IEE132 pKa = 4.2IQCNTGMPWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTMKK151 pKa = 10.32GVSLVPAATGSGTQFSDD168 pKa = 4.17FLEE171 pKa = 4.5TSNGYY176 pKa = 8.07MRR178 pKa = 11.84VMNQVNGNPSLDD190 pKa = 3.28PMYY193 pKa = 11.03SLFGALFKK201 pKa = 11.24GQVNSDD207 pKa = 2.95WQDD210 pKa = 2.91PMTAPTDD217 pKa = 3.57NSRR220 pKa = 11.84LSIKK224 pKa = 9.81YY225 pKa = 10.43DD226 pKa = 3.17KK227 pKa = 10.17VTSLASGNEE236 pKa = 3.89DD237 pKa = 3.44GFIRR241 pKa = 11.84SYY243 pKa = 11.1KK244 pKa = 9.57RR245 pKa = 11.84WHH247 pKa = 6.77PMNKK251 pKa = 8.21TLVYY255 pKa = 10.82NDD257 pKa = 4.58DD258 pKa = 3.83EE259 pKa = 5.64LGGGMLSNFKK269 pKa = 10.9SSFGKK274 pKa = 10.48AGMGDD279 pKa = 4.22YY280 pKa = 10.8YY281 pKa = 11.48VVDD284 pKa = 3.75IFRR287 pKa = 11.84ARR289 pKa = 11.84QGSATTDD296 pKa = 3.36QLSVCPEE303 pKa = 3.55ATLYY307 pKa = 9.32WHH309 pKa = 7.08EE310 pKa = 4.26KK311 pKa = 9.3
Molecular weight: 34.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
618 |
307 |
311 |
309.0 |
34.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.9 ± 0.075 | 1.942 ± 0.009 |
5.987 ± 1.077 | 3.722 ± 1.066 |
5.663 ± 1.774 | 8.576 ± 0.077 |
2.104 ± 1.059 | 3.56 ± 0.716 |
5.502 ± 1.373 | 5.987 ± 0.378 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.074 ± 0.801 | 4.207 ± 0.447 |
4.369 ± 0.603 | 2.589 ± 0.454 |
6.472 ± 0.203 | 7.443 ± 1.597 |
6.796 ± 2.299 | 6.311 ± 1.078 |
2.427 ± 0.128 | 4.369 ± 0.562 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
