
Massariosphaeria phaeospora
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Pleosporomycetidae; Pleosporales; Pleosporales incertae sedis; Massariosphaeria
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
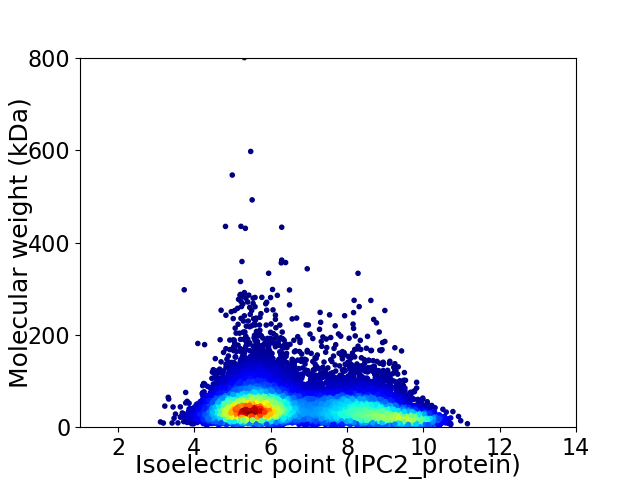
Virtual 2D-PAGE plot for 14012 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7C8I250|A0A7C8I250_9PLEO Histidine biosynthesis trifunctional protein OS=Massariosphaeria phaeospora OX=100035 GN=BDV95DRAFT_525914 PE=3 SV=1
MM1 pKa = 7.42ARR3 pKa = 11.84EE4 pKa = 4.48CVSCLSPEE12 pKa = 4.19EE13 pKa = 4.22EE14 pKa = 4.4CPLRR18 pKa = 11.84QTPCGDD24 pKa = 3.44HH25 pKa = 6.13YY26 pKa = 11.73LCDD29 pKa = 3.59EE30 pKa = 4.54CLEE33 pKa = 4.08QLFTLALNDD42 pKa = 3.66EE43 pKa = 4.25KK44 pKa = 10.83HH45 pKa = 5.71YY46 pKa = 10.58PPRR49 pKa = 11.84CCRR52 pKa = 11.84HH53 pKa = 5.09QSSLLLIGEE62 pKa = 4.77FEE64 pKa = 4.15QHH66 pKa = 6.61LPAQLMVEE74 pKa = 4.23YY75 pKa = 9.88QAKK78 pKa = 10.3EE79 pKa = 3.77NGEE82 pKa = 4.17YY83 pKa = 10.47AVQPRR88 pKa = 11.84FRR90 pKa = 11.84RR91 pKa = 11.84YY92 pKa = 9.86CADD95 pKa = 3.65PSCAKK100 pKa = 10.19FLRR103 pKa = 11.84PASHH107 pKa = 6.43ITDD110 pKa = 3.02SDD112 pKa = 3.75MNITYY117 pKa = 9.46AICEE121 pKa = 3.84ADD123 pKa = 3.44KK124 pKa = 10.92CLKK127 pKa = 8.79ATCIDD132 pKa = 3.93CRR134 pKa = 11.84TLLLNGTADD143 pKa = 3.81HH144 pKa = 6.16VCEE147 pKa = 5.19VPDD150 pKa = 3.5SDD152 pKa = 5.28KK153 pKa = 11.25KK154 pKa = 10.92FKK156 pKa = 9.58EE157 pKa = 3.95TAKK160 pKa = 10.42EE161 pKa = 4.0KK162 pKa = 10.61GYY164 pKa = 10.1QSCFACGSTVEE175 pKa = 4.2LAEE178 pKa = 4.3ACNHH182 pKa = 5.11ITCGCGNNFCYY193 pKa = 10.75VCGKK197 pKa = 8.92EE198 pKa = 3.83WPGMHH203 pKa = 7.14GCPQYY208 pKa = 11.39GPAVYY213 pKa = 10.31DD214 pKa = 3.57EE215 pKa = 4.68EE216 pKa = 6.46GYY218 pKa = 10.22NQDD221 pKa = 4.72GYY223 pKa = 11.63HH224 pKa = 7.11KK225 pKa = 10.13DD226 pKa = 2.99TGLNRR231 pKa = 11.84DD232 pKa = 3.07GLTRR236 pKa = 11.84VQVHH240 pKa = 5.84IRR242 pKa = 11.84DD243 pKa = 3.95GADD246 pKa = 3.65DD247 pKa = 6.15DD248 pKa = 5.99DD249 pKa = 7.39DD250 pKa = 5.72EE251 pKa = 7.9DD252 pKa = 6.76DD253 pKa = 5.76DD254 pKa = 4.86VDD256 pKa = 4.41ANRR259 pKa = 11.84DD260 pKa = 3.54EE261 pKa = 6.33DD262 pKa = 5.01DD263 pKa = 4.06EE264 pKa = 5.66DD265 pKa = 3.57WDD267 pKa = 4.36EE268 pKa = 5.07EE269 pKa = 4.57VLQHH273 pKa = 7.49LDD275 pKa = 3.18DD276 pKa = 4.93DD277 pKa = 4.09QRR279 pKa = 11.84AMINNMDD286 pKa = 3.75PVLRR290 pKa = 11.84EE291 pKa = 3.48EE292 pKa = 4.07TLEE295 pKa = 4.01VFRR298 pKa = 11.84VQLFEE303 pKa = 3.79EE304 pKa = 4.42RR305 pKa = 11.84GIIFQQGDD313 pKa = 3.66GHH315 pKa = 6.26HH316 pKa = 6.4HH317 pKa = 6.12HH318 pKa = 6.43QHH320 pKa = 5.55GQDD323 pKa = 3.38LARR326 pKa = 11.84EE327 pKa = 4.2DD328 pKa = 5.45DD329 pKa = 4.69ADD331 pKa = 6.19DD332 pKa = 6.01DD333 pKa = 6.27DD334 pKa = 6.76ADD336 pKa = 3.66QDD338 pKa = 4.03NPSDD342 pKa = 4.21EE343 pKa = 5.8DD344 pKa = 5.28DD345 pKa = 6.23GDD347 pKa = 4.85DD348 pKa = 5.25QDD350 pKa = 6.33DD351 pKa = 4.65DD352 pKa = 5.01QDD354 pKa = 5.42DD355 pKa = 4.62DD356 pKa = 5.54GNDD359 pKa = 4.21DD360 pKa = 4.02EE361 pKa = 6.72LEE363 pKa = 4.54DD364 pKa = 3.89PTDD367 pKa = 3.32EE368 pKa = 5.52RR369 pKa = 11.84FADD372 pKa = 3.97DD373 pKa = 3.62QSADD377 pKa = 3.49EE378 pKa = 4.84ALFDD382 pKa = 4.53IDD384 pKa = 5.98DD385 pKa = 3.84IVASVEE391 pKa = 4.28DD392 pKa = 3.92VLATADD398 pKa = 4.39AINDD402 pKa = 3.36IVSGVEE408 pKa = 3.52AGLATTSQDD417 pKa = 5.1DD418 pKa = 4.2EE419 pKa = 5.55IEE421 pKa = 4.58DD422 pKa = 3.59EE423 pKa = 4.54GDD425 pKa = 3.71ALVHH429 pKa = 6.27SPADD433 pKa = 3.8SAILPALSANEE444 pKa = 3.79DD445 pKa = 4.15DD446 pKa = 4.39AWSSNSPVSPPQQDD460 pKa = 3.2LTEE463 pKa = 3.99QSIEE467 pKa = 4.12PKK469 pKa = 10.23TPGDD473 pKa = 3.64VMQLSDD479 pKa = 4.75AAYY482 pKa = 10.4GDD484 pKa = 4.47DD485 pKa = 4.46NGGAGAGPMYY495 pKa = 10.76GGGLTGWGGGEE506 pKa = 4.0EE507 pKa = 4.17LL508 pKa = 4.88
MM1 pKa = 7.42ARR3 pKa = 11.84EE4 pKa = 4.48CVSCLSPEE12 pKa = 4.19EE13 pKa = 4.22EE14 pKa = 4.4CPLRR18 pKa = 11.84QTPCGDD24 pKa = 3.44HH25 pKa = 6.13YY26 pKa = 11.73LCDD29 pKa = 3.59EE30 pKa = 4.54CLEE33 pKa = 4.08QLFTLALNDD42 pKa = 3.66EE43 pKa = 4.25KK44 pKa = 10.83HH45 pKa = 5.71YY46 pKa = 10.58PPRR49 pKa = 11.84CCRR52 pKa = 11.84HH53 pKa = 5.09QSSLLLIGEE62 pKa = 4.77FEE64 pKa = 4.15QHH66 pKa = 6.61LPAQLMVEE74 pKa = 4.23YY75 pKa = 9.88QAKK78 pKa = 10.3EE79 pKa = 3.77NGEE82 pKa = 4.17YY83 pKa = 10.47AVQPRR88 pKa = 11.84FRR90 pKa = 11.84RR91 pKa = 11.84YY92 pKa = 9.86CADD95 pKa = 3.65PSCAKK100 pKa = 10.19FLRR103 pKa = 11.84PASHH107 pKa = 6.43ITDD110 pKa = 3.02SDD112 pKa = 3.75MNITYY117 pKa = 9.46AICEE121 pKa = 3.84ADD123 pKa = 3.44KK124 pKa = 10.92CLKK127 pKa = 8.79ATCIDD132 pKa = 3.93CRR134 pKa = 11.84TLLLNGTADD143 pKa = 3.81HH144 pKa = 6.16VCEE147 pKa = 5.19VPDD150 pKa = 3.5SDD152 pKa = 5.28KK153 pKa = 11.25KK154 pKa = 10.92FKK156 pKa = 9.58EE157 pKa = 3.95TAKK160 pKa = 10.42EE161 pKa = 4.0KK162 pKa = 10.61GYY164 pKa = 10.1QSCFACGSTVEE175 pKa = 4.2LAEE178 pKa = 4.3ACNHH182 pKa = 5.11ITCGCGNNFCYY193 pKa = 10.75VCGKK197 pKa = 8.92EE198 pKa = 3.83WPGMHH203 pKa = 7.14GCPQYY208 pKa = 11.39GPAVYY213 pKa = 10.31DD214 pKa = 3.57EE215 pKa = 4.68EE216 pKa = 6.46GYY218 pKa = 10.22NQDD221 pKa = 4.72GYY223 pKa = 11.63HH224 pKa = 7.11KK225 pKa = 10.13DD226 pKa = 2.99TGLNRR231 pKa = 11.84DD232 pKa = 3.07GLTRR236 pKa = 11.84VQVHH240 pKa = 5.84IRR242 pKa = 11.84DD243 pKa = 3.95GADD246 pKa = 3.65DD247 pKa = 6.15DD248 pKa = 5.99DD249 pKa = 7.39DD250 pKa = 5.72EE251 pKa = 7.9DD252 pKa = 6.76DD253 pKa = 5.76DD254 pKa = 4.86VDD256 pKa = 4.41ANRR259 pKa = 11.84DD260 pKa = 3.54EE261 pKa = 6.33DD262 pKa = 5.01DD263 pKa = 4.06EE264 pKa = 5.66DD265 pKa = 3.57WDD267 pKa = 4.36EE268 pKa = 5.07EE269 pKa = 4.57VLQHH273 pKa = 7.49LDD275 pKa = 3.18DD276 pKa = 4.93DD277 pKa = 4.09QRR279 pKa = 11.84AMINNMDD286 pKa = 3.75PVLRR290 pKa = 11.84EE291 pKa = 3.48EE292 pKa = 4.07TLEE295 pKa = 4.01VFRR298 pKa = 11.84VQLFEE303 pKa = 3.79EE304 pKa = 4.42RR305 pKa = 11.84GIIFQQGDD313 pKa = 3.66GHH315 pKa = 6.26HH316 pKa = 6.4HH317 pKa = 6.12HH318 pKa = 6.43QHH320 pKa = 5.55GQDD323 pKa = 3.38LARR326 pKa = 11.84EE327 pKa = 4.2DD328 pKa = 5.45DD329 pKa = 4.69ADD331 pKa = 6.19DD332 pKa = 6.01DD333 pKa = 6.27DD334 pKa = 6.76ADD336 pKa = 3.66QDD338 pKa = 4.03NPSDD342 pKa = 4.21EE343 pKa = 5.8DD344 pKa = 5.28DD345 pKa = 6.23GDD347 pKa = 4.85DD348 pKa = 5.25QDD350 pKa = 6.33DD351 pKa = 4.65DD352 pKa = 5.01QDD354 pKa = 5.42DD355 pKa = 4.62DD356 pKa = 5.54GNDD359 pKa = 4.21DD360 pKa = 4.02EE361 pKa = 6.72LEE363 pKa = 4.54DD364 pKa = 3.89PTDD367 pKa = 3.32EE368 pKa = 5.52RR369 pKa = 11.84FADD372 pKa = 3.97DD373 pKa = 3.62QSADD377 pKa = 3.49EE378 pKa = 4.84ALFDD382 pKa = 4.53IDD384 pKa = 5.98DD385 pKa = 3.84IVASVEE391 pKa = 4.28DD392 pKa = 3.92VLATADD398 pKa = 4.39AINDD402 pKa = 3.36IVSGVEE408 pKa = 3.52AGLATTSQDD417 pKa = 5.1DD418 pKa = 4.2EE419 pKa = 5.55IEE421 pKa = 4.58DD422 pKa = 3.59EE423 pKa = 4.54GDD425 pKa = 3.71ALVHH429 pKa = 6.27SPADD433 pKa = 3.8SAILPALSANEE444 pKa = 3.79DD445 pKa = 4.15DD446 pKa = 4.39AWSSNSPVSPPQQDD460 pKa = 3.2LTEE463 pKa = 3.99QSIEE467 pKa = 4.12PKK469 pKa = 10.23TPGDD473 pKa = 3.64VMQLSDD479 pKa = 4.75AAYY482 pKa = 10.4GDD484 pKa = 4.47DD485 pKa = 4.46NGGAGAGPMYY495 pKa = 10.76GGGLTGWGGGEE506 pKa = 4.0EE507 pKa = 4.17LL508 pKa = 4.88
Molecular weight: 56.03 kDa
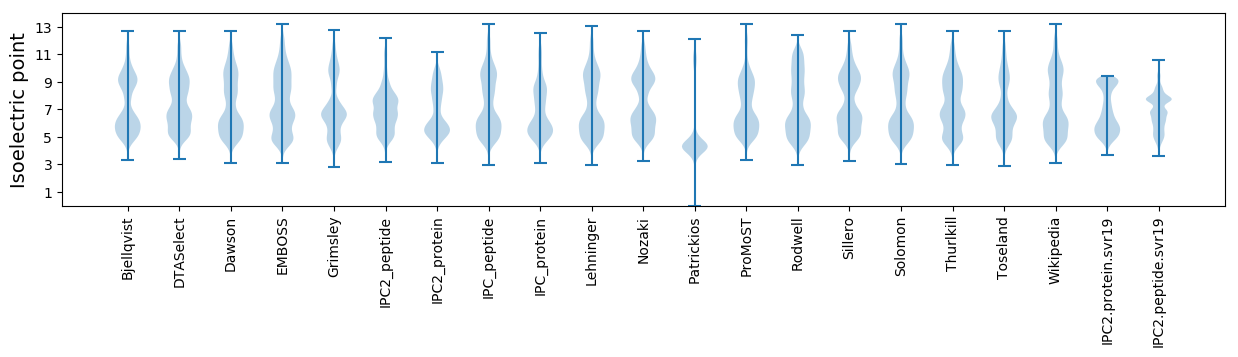
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7C8I4H0|A0A7C8I4H0_9PLEO Uncharacterized protein OS=Massariosphaeria phaeospora OX=100035 GN=BDV95DRAFT_167222 PE=4 SV=1
MM1 pKa = 7.23NALRR5 pKa = 11.84QHH7 pKa = 5.91SNRR10 pKa = 11.84PQSHH14 pKa = 6.05KK15 pKa = 10.84SFRR18 pKa = 11.84TKK20 pKa = 10.44QKK22 pKa = 10.05LARR25 pKa = 11.84AQKK28 pKa = 8.97QNRR31 pKa = 11.84PIPQWIRR38 pKa = 11.84LRR40 pKa = 11.84TNNTIRR46 pKa = 11.84YY47 pKa = 5.73NAKK50 pKa = 8.89RR51 pKa = 11.84RR52 pKa = 11.84HH53 pKa = 4.14WRR55 pKa = 11.84KK56 pKa = 7.41TRR58 pKa = 11.84LGII61 pKa = 4.46
MM1 pKa = 7.23NALRR5 pKa = 11.84QHH7 pKa = 5.91SNRR10 pKa = 11.84PQSHH14 pKa = 6.05KK15 pKa = 10.84SFRR18 pKa = 11.84TKK20 pKa = 10.44QKK22 pKa = 10.05LARR25 pKa = 11.84AQKK28 pKa = 8.97QNRR31 pKa = 11.84PIPQWIRR38 pKa = 11.84LRR40 pKa = 11.84TNNTIRR46 pKa = 11.84YY47 pKa = 5.73NAKK50 pKa = 8.89RR51 pKa = 11.84RR52 pKa = 11.84HH53 pKa = 4.14WRR55 pKa = 11.84KK56 pKa = 7.41TRR58 pKa = 11.84LGII61 pKa = 4.46
Molecular weight: 7.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6065376 |
49 |
7228 |
432.9 |
47.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.882 ± 0.019 | 1.322 ± 0.01 |
5.619 ± 0.016 | 6.003 ± 0.019 |
3.647 ± 0.013 | 6.773 ± 0.018 |
2.563 ± 0.01 | 4.62 ± 0.014 |
4.773 ± 0.021 | 8.893 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.154 ± 0.007 | 3.546 ± 0.013 |
6.335 ± 0.024 | 3.996 ± 0.013 |
6.358 ± 0.021 | 8.039 ± 0.024 |
6.114 ± 0.013 | 6.123 ± 0.014 |
1.499 ± 0.008 | 2.742 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
