
Rathayibacter sp. Leaf296
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Rathayibacter; unclassified Rathayibacter
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
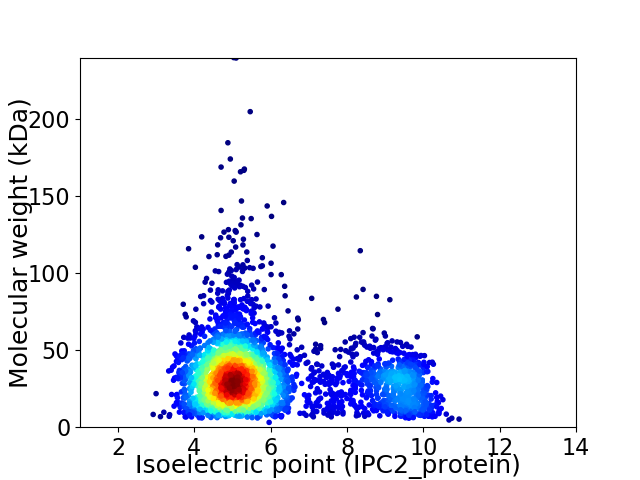
Virtual 2D-PAGE plot for 3537 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5B832|A0A0Q5B832_9MICO Acetyltransferase OS=Rathayibacter sp. Leaf296 OX=1736327 GN=ASF46_13405 PE=4 SV=1
MM1 pKa = 7.8KK2 pKa = 9.97IRR4 pKa = 11.84RR5 pKa = 11.84FGLGALALVAASTLVLAGCSSSNDD29 pKa = 3.69DD30 pKa = 4.35GGGDD34 pKa = 3.62AAGDD38 pKa = 3.67SAAIILTNGSEE49 pKa = 4.23PQNPLVPTNTNEE61 pKa = 3.92VGGGKK66 pKa = 9.3ILDD69 pKa = 4.15EE70 pKa = 4.46IFAGLYY76 pKa = 10.44YY77 pKa = 10.61YY78 pKa = 10.52DD79 pKa = 4.87AEE81 pKa = 4.68GAAQPDD87 pKa = 3.76LAEE90 pKa = 4.45SVEE93 pKa = 4.63SDD95 pKa = 3.18DD96 pKa = 3.87AQNYY100 pKa = 7.27TITIEE105 pKa = 4.06SGRR108 pKa = 11.84TFTNGEE114 pKa = 4.31EE115 pKa = 3.71ITAQSFVDD123 pKa = 3.45AWNYY127 pKa = 7.51GAKK130 pKa = 10.22YY131 pKa = 10.63SNEE134 pKa = 3.64QLQSYY139 pKa = 8.54FFEE142 pKa = 6.24DD143 pKa = 3.26IEE145 pKa = 4.47GFSYY149 pKa = 11.31DD150 pKa = 4.23EE151 pKa = 4.3DD152 pKa = 4.4TEE154 pKa = 4.38LTGLKK159 pKa = 10.31VVDD162 pKa = 3.79DD163 pKa = 4.05TTFTVALTAPKK174 pKa = 10.33SDD176 pKa = 3.26FPQRR180 pKa = 11.84LGYY183 pKa = 8.89SAYY186 pKa = 10.6YY187 pKa = 8.77PLPSVAYY194 pKa = 10.2DD195 pKa = 3.51DD196 pKa = 3.86MDD198 pKa = 4.43AFGEE202 pKa = 4.15NPIGNGPYY210 pKa = 9.65MLDD213 pKa = 4.94GEE215 pKa = 5.11GAWQHH220 pKa = 6.42DD221 pKa = 4.06VQIDD225 pKa = 3.87LVVNPDD231 pKa = 3.1YY232 pKa = 11.1DD233 pKa = 3.98GPRR236 pKa = 11.84VPVNGGITITFYY248 pKa = 10.33ATQDD252 pKa = 2.86ASYY255 pKa = 11.65ADD257 pKa = 4.25LLAGNVDD264 pKa = 4.58VIDD267 pKa = 4.71QIPDD271 pKa = 3.29SAFGTFEE278 pKa = 4.19SDD280 pKa = 3.4LGDD283 pKa = 3.4RR284 pKa = 11.84AVNQASAVFQSFTIPEE300 pKa = 4.15RR301 pKa = 11.84LEE303 pKa = 4.16HH304 pKa = 6.82FSGEE308 pKa = 4.03EE309 pKa = 3.73GKK311 pKa = 10.49LRR313 pKa = 11.84RR314 pKa = 11.84QAISMAFDD322 pKa = 3.58RR323 pKa = 11.84EE324 pKa = 4.2QITDD328 pKa = 3.84VIFEE332 pKa = 4.27GTRR335 pKa = 11.84TPASDD340 pKa = 3.36FTSPVIDD347 pKa = 3.67GWSDD351 pKa = 3.32SLEE354 pKa = 4.07GAEE357 pKa = 3.84VLEE360 pKa = 4.3YY361 pKa = 11.26NPDD364 pKa = 3.47EE365 pKa = 4.61AKK367 pKa = 10.84KK368 pKa = 10.38LWAEE372 pKa = 4.09ADD374 pKa = 4.46AISPWTGTFSIAYY387 pKa = 8.93NADD390 pKa = 3.37GGHH393 pKa = 5.87QGWVDD398 pKa = 3.36AVSNAVKK405 pKa = 9.3NTLGIDD411 pKa = 3.87AVGEE415 pKa = 4.08AYY417 pKa = 7.62PTFAEE422 pKa = 4.12ARR424 pKa = 11.84TKK426 pKa = 10.45IVDD429 pKa = 3.65RR430 pKa = 11.84SIEE433 pKa = 4.09TAFRR437 pKa = 11.84TGWQADD443 pKa = 3.65YY444 pKa = 10.71PGLYY448 pKa = 10.33NFLGPLYY455 pKa = 9.65ATNASSNDD463 pKa = 3.02GDD465 pKa = 3.9YY466 pKa = 11.5SNPEE470 pKa = 3.85FDD472 pKa = 4.7QLLADD477 pKa = 5.07GSAEE481 pKa = 4.04TDD483 pKa = 3.15LATANEE489 pKa = 5.01DD490 pKa = 3.44YY491 pKa = 11.16QKK493 pKa = 11.05AQEE496 pKa = 4.51VLLQDD501 pKa = 4.65LPAIPLWYY509 pKa = 10.55ANVTGGYY516 pKa = 9.21SDD518 pKa = 3.73TVSDD522 pKa = 6.05VEE524 pKa = 4.56FGWNSVPLFYY534 pKa = 10.77QITKK538 pKa = 10.24SS539 pKa = 3.29
MM1 pKa = 7.8KK2 pKa = 9.97IRR4 pKa = 11.84RR5 pKa = 11.84FGLGALALVAASTLVLAGCSSSNDD29 pKa = 3.69DD30 pKa = 4.35GGGDD34 pKa = 3.62AAGDD38 pKa = 3.67SAAIILTNGSEE49 pKa = 4.23PQNPLVPTNTNEE61 pKa = 3.92VGGGKK66 pKa = 9.3ILDD69 pKa = 4.15EE70 pKa = 4.46IFAGLYY76 pKa = 10.44YY77 pKa = 10.61YY78 pKa = 10.52DD79 pKa = 4.87AEE81 pKa = 4.68GAAQPDD87 pKa = 3.76LAEE90 pKa = 4.45SVEE93 pKa = 4.63SDD95 pKa = 3.18DD96 pKa = 3.87AQNYY100 pKa = 7.27TITIEE105 pKa = 4.06SGRR108 pKa = 11.84TFTNGEE114 pKa = 4.31EE115 pKa = 3.71ITAQSFVDD123 pKa = 3.45AWNYY127 pKa = 7.51GAKK130 pKa = 10.22YY131 pKa = 10.63SNEE134 pKa = 3.64QLQSYY139 pKa = 8.54FFEE142 pKa = 6.24DD143 pKa = 3.26IEE145 pKa = 4.47GFSYY149 pKa = 11.31DD150 pKa = 4.23EE151 pKa = 4.3DD152 pKa = 4.4TEE154 pKa = 4.38LTGLKK159 pKa = 10.31VVDD162 pKa = 3.79DD163 pKa = 4.05TTFTVALTAPKK174 pKa = 10.33SDD176 pKa = 3.26FPQRR180 pKa = 11.84LGYY183 pKa = 8.89SAYY186 pKa = 10.6YY187 pKa = 8.77PLPSVAYY194 pKa = 10.2DD195 pKa = 3.51DD196 pKa = 3.86MDD198 pKa = 4.43AFGEE202 pKa = 4.15NPIGNGPYY210 pKa = 9.65MLDD213 pKa = 4.94GEE215 pKa = 5.11GAWQHH220 pKa = 6.42DD221 pKa = 4.06VQIDD225 pKa = 3.87LVVNPDD231 pKa = 3.1YY232 pKa = 11.1DD233 pKa = 3.98GPRR236 pKa = 11.84VPVNGGITITFYY248 pKa = 10.33ATQDD252 pKa = 2.86ASYY255 pKa = 11.65ADD257 pKa = 4.25LLAGNVDD264 pKa = 4.58VIDD267 pKa = 4.71QIPDD271 pKa = 3.29SAFGTFEE278 pKa = 4.19SDD280 pKa = 3.4LGDD283 pKa = 3.4RR284 pKa = 11.84AVNQASAVFQSFTIPEE300 pKa = 4.15RR301 pKa = 11.84LEE303 pKa = 4.16HH304 pKa = 6.82FSGEE308 pKa = 4.03EE309 pKa = 3.73GKK311 pKa = 10.49LRR313 pKa = 11.84RR314 pKa = 11.84QAISMAFDD322 pKa = 3.58RR323 pKa = 11.84EE324 pKa = 4.2QITDD328 pKa = 3.84VIFEE332 pKa = 4.27GTRR335 pKa = 11.84TPASDD340 pKa = 3.36FTSPVIDD347 pKa = 3.67GWSDD351 pKa = 3.32SLEE354 pKa = 4.07GAEE357 pKa = 3.84VLEE360 pKa = 4.3YY361 pKa = 11.26NPDD364 pKa = 3.47EE365 pKa = 4.61AKK367 pKa = 10.84KK368 pKa = 10.38LWAEE372 pKa = 4.09ADD374 pKa = 4.46AISPWTGTFSIAYY387 pKa = 8.93NADD390 pKa = 3.37GGHH393 pKa = 5.87QGWVDD398 pKa = 3.36AVSNAVKK405 pKa = 9.3NTLGIDD411 pKa = 3.87AVGEE415 pKa = 4.08AYY417 pKa = 7.62PTFAEE422 pKa = 4.12ARR424 pKa = 11.84TKK426 pKa = 10.45IVDD429 pKa = 3.65RR430 pKa = 11.84SIEE433 pKa = 4.09TAFRR437 pKa = 11.84TGWQADD443 pKa = 3.65YY444 pKa = 10.71PGLYY448 pKa = 10.33NFLGPLYY455 pKa = 9.65ATNASSNDD463 pKa = 3.02GDD465 pKa = 3.9YY466 pKa = 11.5SNPEE470 pKa = 3.85FDD472 pKa = 4.7QLLADD477 pKa = 5.07GSAEE481 pKa = 4.04TDD483 pKa = 3.15LATANEE489 pKa = 5.01DD490 pKa = 3.44YY491 pKa = 11.16QKK493 pKa = 11.05AQEE496 pKa = 4.51VLLQDD501 pKa = 4.65LPAIPLWYY509 pKa = 10.55ANVTGGYY516 pKa = 9.21SDD518 pKa = 3.73TVSDD522 pKa = 6.05VEE524 pKa = 4.56FGWNSVPLFYY534 pKa = 10.77QITKK538 pKa = 10.24SS539 pKa = 3.29
Molecular weight: 58.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q5B367|A0A0Q5B367_9MICO Release factor glutamine methyltransferase OS=Rathayibacter sp. Leaf296 OX=1736327 GN=prmC PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84GILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.56GRR40 pKa = 11.84TEE42 pKa = 4.14LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84GILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.56GRR40 pKa = 11.84TEE42 pKa = 4.14LSAA45 pKa = 4.86
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1140147 |
29 |
2420 |
322.3 |
34.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.697 ± 0.061 | 0.483 ± 0.009 |
6.104 ± 0.039 | 5.885 ± 0.047 |
3.028 ± 0.022 | 9.09 ± 0.036 |
1.844 ± 0.019 | 4.044 ± 0.033 |
1.615 ± 0.029 | 10.717 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.502 ± 0.016 | 1.69 ± 0.02 |
5.484 ± 0.029 | 2.472 ± 0.023 |
7.55 ± 0.05 | 6.078 ± 0.031 |
6.084 ± 0.046 | 9.287 ± 0.039 |
1.424 ± 0.019 | 1.921 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
