
Nocardioides dokdonensis FR1436
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; Nocardioides dokdonensis
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
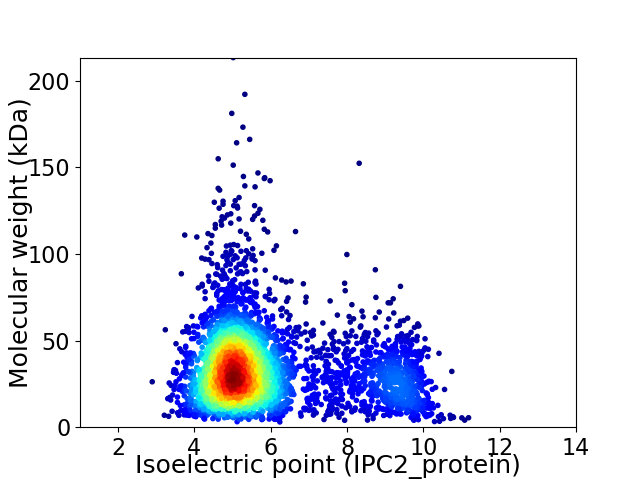
Virtual 2D-PAGE plot for 4089 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A9GP40|A0A1A9GP40_9ACTN Uncharacterized protein OS=Nocardioides dokdonensis FR1436 OX=1300347 GN=I601_2789 PE=4 SV=1
MM1 pKa = 7.8AGTAALGLVLTACGSEE17 pKa = 4.02DD18 pKa = 3.99SEE20 pKa = 5.45PEE22 pKa = 3.78ATDD25 pKa = 3.61NEE27 pKa = 4.58SSSEE31 pKa = 4.29STDD34 pKa = 3.2SAAPADD40 pKa = 4.39DD41 pKa = 4.68EE42 pKa = 4.69FTNDD46 pKa = 3.17EE47 pKa = 4.68CPNGSTSDD55 pKa = 4.0DD56 pKa = 3.61SFKK59 pKa = 11.28AGGILPLTGNLAFLGPPEE77 pKa = 4.02VAGVGMAVSDD87 pKa = 3.58INAAGGVNGADD98 pKa = 3.14ACHH101 pKa = 6.06QIEE104 pKa = 4.89DD105 pKa = 4.3SGDD108 pKa = 3.57TTDD111 pKa = 5.52LSISTASARR120 pKa = 11.84KK121 pKa = 9.63LIQDD125 pKa = 3.61KK126 pKa = 10.98ASVVIGAASSSVSLNFVDD144 pKa = 5.07RR145 pKa = 11.84LTAAKK150 pKa = 8.51ITQVSPANTAVDD162 pKa = 3.86LSGYY166 pKa = 9.7SDD168 pKa = 4.08FYY170 pKa = 11.37FRR172 pKa = 11.84TAPPDD177 pKa = 4.67GIQGNALGSLISSDD191 pKa = 3.61GFQKK195 pKa = 10.6VAFIVFSDD203 pKa = 3.98TYY205 pKa = 10.01GTGLRR210 pKa = 11.84DD211 pKa = 3.58VVQEE215 pKa = 4.16TIEE218 pKa = 4.32AGGGEE223 pKa = 4.42CTYY226 pKa = 11.01GCKK229 pKa = 10.21GDD231 pKa = 4.13GDD233 pKa = 4.34EE234 pKa = 5.39FPAGQTTFSSEE245 pKa = 3.43VSAATNSNPDD255 pKa = 3.92AIVILAFDD263 pKa = 3.78EE264 pKa = 4.52TKK266 pKa = 10.82AIVPEE271 pKa = 4.19LASTGWDD278 pKa = 3.06MSATYY283 pKa = 10.24FSDD286 pKa = 3.68GNTADD291 pKa = 3.62FSEE294 pKa = 4.76DD295 pKa = 3.37FEE297 pKa = 6.93AGTLTGAQGTIPGADD312 pKa = 3.09AAQDD316 pKa = 3.67FKK318 pKa = 11.8DD319 pKa = 5.38RR320 pKa = 11.84LVAWNEE326 pKa = 3.61YY327 pKa = 10.3AFNDD331 pKa = 3.75GLTDD335 pKa = 3.71FAYY338 pKa = 10.21AAEE341 pKa = 4.85SYY343 pKa = 10.73DD344 pKa = 3.64ATILAALAAYY354 pKa = 9.52KK355 pKa = 10.66GGATDD360 pKa = 3.88SVTVQKK366 pKa = 11.1NFAAVSGATGGEE378 pKa = 4.29EE379 pKa = 4.08CTSYY383 pKa = 11.69ADD385 pKa = 3.57CTAMLDD391 pKa = 3.41EE392 pKa = 5.03GNEE395 pKa = 3.67IRR397 pKa = 11.84YY398 pKa = 8.93VGPSGIGPIDD408 pKa = 3.94EE409 pKa = 5.0EE410 pKa = 4.57NDD412 pKa = 3.35PSSAFVGIYY421 pKa = 9.35TYY423 pKa = 11.4NEE425 pKa = 3.7NNKK428 pKa = 9.75NEE430 pKa = 4.11LTTTVEE436 pKa = 4.3GQKK439 pKa = 10.43QSS441 pKa = 3.09
MM1 pKa = 7.8AGTAALGLVLTACGSEE17 pKa = 4.02DD18 pKa = 3.99SEE20 pKa = 5.45PEE22 pKa = 3.78ATDD25 pKa = 3.61NEE27 pKa = 4.58SSSEE31 pKa = 4.29STDD34 pKa = 3.2SAAPADD40 pKa = 4.39DD41 pKa = 4.68EE42 pKa = 4.69FTNDD46 pKa = 3.17EE47 pKa = 4.68CPNGSTSDD55 pKa = 4.0DD56 pKa = 3.61SFKK59 pKa = 11.28AGGILPLTGNLAFLGPPEE77 pKa = 4.02VAGVGMAVSDD87 pKa = 3.58INAAGGVNGADD98 pKa = 3.14ACHH101 pKa = 6.06QIEE104 pKa = 4.89DD105 pKa = 4.3SGDD108 pKa = 3.57TTDD111 pKa = 5.52LSISTASARR120 pKa = 11.84KK121 pKa = 9.63LIQDD125 pKa = 3.61KK126 pKa = 10.98ASVVIGAASSSVSLNFVDD144 pKa = 5.07RR145 pKa = 11.84LTAAKK150 pKa = 8.51ITQVSPANTAVDD162 pKa = 3.86LSGYY166 pKa = 9.7SDD168 pKa = 4.08FYY170 pKa = 11.37FRR172 pKa = 11.84TAPPDD177 pKa = 4.67GIQGNALGSLISSDD191 pKa = 3.61GFQKK195 pKa = 10.6VAFIVFSDD203 pKa = 3.98TYY205 pKa = 10.01GTGLRR210 pKa = 11.84DD211 pKa = 3.58VVQEE215 pKa = 4.16TIEE218 pKa = 4.32AGGGEE223 pKa = 4.42CTYY226 pKa = 11.01GCKK229 pKa = 10.21GDD231 pKa = 4.13GDD233 pKa = 4.34EE234 pKa = 5.39FPAGQTTFSSEE245 pKa = 3.43VSAATNSNPDD255 pKa = 3.92AIVILAFDD263 pKa = 3.78EE264 pKa = 4.52TKK266 pKa = 10.82AIVPEE271 pKa = 4.19LASTGWDD278 pKa = 3.06MSATYY283 pKa = 10.24FSDD286 pKa = 3.68GNTADD291 pKa = 3.62FSEE294 pKa = 4.76DD295 pKa = 3.37FEE297 pKa = 6.93AGTLTGAQGTIPGADD312 pKa = 3.09AAQDD316 pKa = 3.67FKK318 pKa = 11.8DD319 pKa = 5.38RR320 pKa = 11.84LVAWNEE326 pKa = 3.61YY327 pKa = 10.3AFNDD331 pKa = 3.75GLTDD335 pKa = 3.71FAYY338 pKa = 10.21AAEE341 pKa = 4.85SYY343 pKa = 10.73DD344 pKa = 3.64ATILAALAAYY354 pKa = 9.52KK355 pKa = 10.66GGATDD360 pKa = 3.88SVTVQKK366 pKa = 11.1NFAAVSGATGGEE378 pKa = 4.29EE379 pKa = 4.08CTSYY383 pKa = 11.69ADD385 pKa = 3.57CTAMLDD391 pKa = 3.41EE392 pKa = 5.03GNEE395 pKa = 3.67IRR397 pKa = 11.84YY398 pKa = 8.93VGPSGIGPIDD408 pKa = 3.94EE409 pKa = 5.0EE410 pKa = 4.57NDD412 pKa = 3.35PSSAFVGIYY421 pKa = 9.35TYY423 pKa = 11.4NEE425 pKa = 3.7NNKK428 pKa = 9.75NEE430 pKa = 4.11LTTTVEE436 pKa = 4.3GQKK439 pKa = 10.43QSS441 pKa = 3.09
Molecular weight: 45.24 kDa
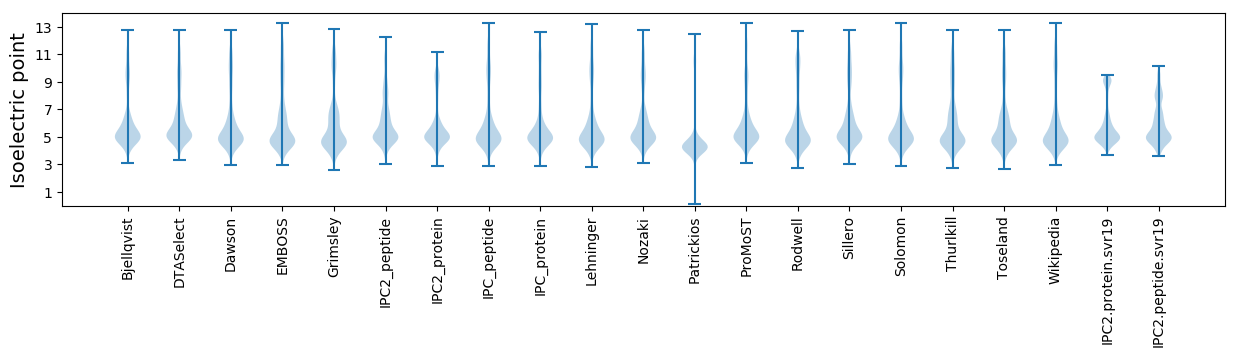
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A9GGE0|A0A1A9GGE0_9ACTN Orotate phosphoribosyltransferase OS=Nocardioides dokdonensis FR1436 OX=1300347 GN=pyrE_1 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.24TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.24TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
Molecular weight: 4.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1337234 |
29 |
1969 |
327.0 |
34.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.109 ± 0.053 | 0.729 ± 0.01 |
6.532 ± 0.028 | 5.9 ± 0.037 |
2.645 ± 0.022 | 9.374 ± 0.035 |
2.286 ± 0.021 | 3.047 ± 0.027 |
1.74 ± 0.031 | 10.549 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.857 ± 0.015 | 1.554 ± 0.018 |
5.724 ± 0.031 | 2.81 ± 0.02 |
7.828 ± 0.042 | 5.292 ± 0.024 |
6.035 ± 0.03 | 9.599 ± 0.037 |
1.523 ± 0.017 | 1.867 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
