
Beihai narna-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
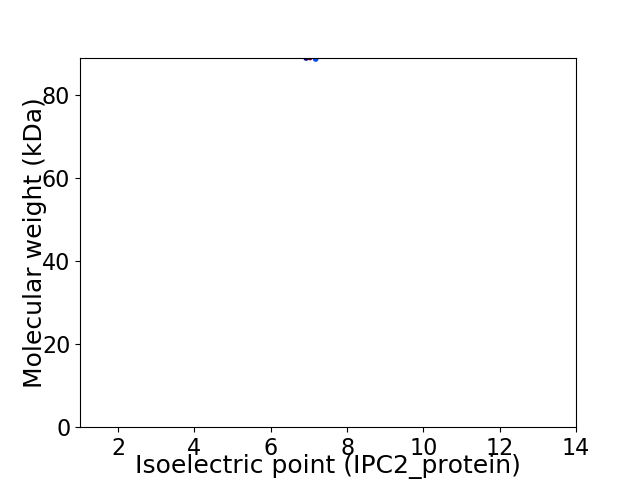
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KI53|A0A1L3KI53_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 1 OX=1922436 PE=4 SV=1
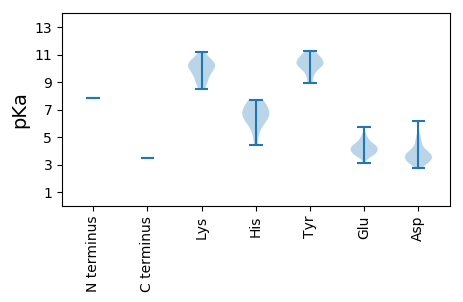
MM1 pKa = 7.86DD2 pKa = 6.2PIEE5 pKa = 4.43VSHH8 pKa = 6.95ALYY11 pKa = 10.52EE12 pKa = 4.88GLASWCMSRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 8.47GTKK26 pKa = 9.76DD27 pKa = 3.18PNFQDD32 pKa = 3.53LCRR35 pKa = 11.84LARR38 pKa = 11.84HH39 pKa = 6.51CGLCEE44 pKa = 3.79PPKK47 pKa = 10.8CLDD50 pKa = 3.34SALTEE55 pKa = 4.28FLNSFDD61 pKa = 4.25PPSCVEE67 pKa = 4.01HH68 pKa = 7.69DD69 pKa = 4.41NILKK73 pKa = 10.02EE74 pKa = 4.39RR75 pKa = 11.84IVHH78 pKa = 7.08SIQQANRR85 pKa = 11.84MVAVYY90 pKa = 10.27TDD92 pKa = 3.03ILKK95 pKa = 10.61FYY97 pKa = 10.36GVLGRR102 pKa = 11.84LHH104 pKa = 6.55PKK106 pKa = 8.68GWLWYY111 pKa = 9.74GVLGLLEE118 pKa = 5.75KK119 pKa = 10.25YY120 pKa = 10.32LKK122 pKa = 10.25WSTADD127 pKa = 3.45FLARR131 pKa = 11.84TLDD134 pKa = 4.33QEE136 pKa = 4.91DD137 pKa = 4.7LPPNPGFEE145 pKa = 4.47LGLPFTGPAASHH157 pKa = 6.83LKK159 pKa = 9.88RR160 pKa = 11.84VCLFNRR166 pKa = 11.84KK167 pKa = 9.18RR168 pKa = 11.84GLRR171 pKa = 11.84STYY174 pKa = 10.76AVALAYY180 pKa = 10.35SLYY183 pKa = 9.5QGKK186 pKa = 10.16GGSLPVSAEE195 pKa = 3.7FLEE198 pKa = 4.54NSVEE202 pKa = 4.06RR203 pKa = 11.84AVNALCTPRR212 pKa = 11.84TGMTEE217 pKa = 4.4DD218 pKa = 3.33IFDD221 pKa = 3.77VMSEE225 pKa = 3.96QVRR228 pKa = 11.84RR229 pKa = 11.84TVRR232 pKa = 11.84EE233 pKa = 3.91CFPRR237 pKa = 11.84HH238 pKa = 4.97HH239 pKa = 6.9SEE241 pKa = 3.69RR242 pKa = 11.84ARR244 pKa = 11.84PGSHH248 pKa = 7.41RR249 pKa = 11.84IPSLRR254 pKa = 11.84SSFQNSRR261 pKa = 11.84KK262 pKa = 9.44DD263 pKa = 3.04WGAFGHH269 pKa = 6.34IVSASISIGFPYY281 pKa = 10.93LLGHH285 pKa = 6.18WHH287 pKa = 6.89HH288 pKa = 6.46KK289 pKa = 8.86TNIEE293 pKa = 3.99LVYY296 pKa = 10.78GPSDD300 pKa = 3.81PEE302 pKa = 4.38DD303 pKa = 3.29VQLDD307 pKa = 3.84YY308 pKa = 10.98EE309 pKa = 4.2QSYY312 pKa = 10.19RR313 pKa = 11.84RR314 pKa = 11.84AVLMGRR320 pKa = 11.84CRR322 pKa = 11.84CYY324 pKa = 10.21PVGLLEE330 pKa = 4.16PFKK333 pKa = 11.21VRR335 pKa = 11.84VITRR339 pKa = 11.84GDD341 pKa = 3.03ADD343 pKa = 3.76MYY345 pKa = 10.73HH346 pKa = 7.53LARR349 pKa = 11.84RR350 pKa = 11.84WQKK353 pKa = 9.5TIHH356 pKa = 5.9GCLRR360 pKa = 11.84QHH362 pKa = 6.69PTFQLIGGPIDD373 pKa = 3.65WSALNHH379 pKa = 5.75LSEE382 pKa = 4.62RR383 pKa = 11.84TDD385 pKa = 3.13ICDD388 pKa = 3.21DD389 pKa = 4.26RR390 pKa = 11.84YY391 pKa = 10.03WVSADD396 pKa = 3.41YY397 pKa = 10.41TSATDD402 pKa = 3.75NLDD405 pKa = 3.75PEE407 pKa = 4.66LSRR410 pKa = 11.84VCMEE414 pKa = 5.24EE415 pKa = 3.11ICQRR419 pKa = 11.84LGVPLEE425 pKa = 4.37DD426 pKa = 3.34QLVLLRR432 pKa = 11.84TLTDD436 pKa = 3.13HH437 pKa = 7.68DD438 pKa = 5.11LYY440 pKa = 11.24EE441 pKa = 4.39EE442 pKa = 4.31SGDD445 pKa = 4.88LIGSQAWGQLMGSPSSFPILCLINAAATRR474 pKa = 11.84FALEE478 pKa = 3.7LANWKK483 pKa = 9.96VGEE486 pKa = 4.27YY487 pKa = 10.79KK488 pKa = 10.18LTDD491 pKa = 3.59LPMLFNGDD499 pKa = 3.55DD500 pKa = 3.32AAFRR504 pKa = 11.84ANEE507 pKa = 3.8QEE509 pKa = 3.88YY510 pKa = 8.91TIWKK514 pKa = 9.71GITKK518 pKa = 10.2SVGLEE523 pKa = 3.54FSVGKK528 pKa = 10.31NFFHH532 pKa = 7.47KK533 pKa = 10.45RR534 pKa = 11.84FLILNSAVHH543 pKa = 6.39EE544 pKa = 4.28DD545 pKa = 3.34VGRR548 pKa = 11.84VDD550 pKa = 3.62FFGRR554 pKa = 11.84RR555 pKa = 11.84FSRR558 pKa = 11.84LEE560 pKa = 3.7PVVTLSSSLLYY571 pKa = 10.65GQVRR575 pKa = 11.84VQSASQNDD583 pKa = 3.92RR584 pKa = 11.84THH586 pKa = 7.16LCQSDD591 pKa = 3.88LSVGRR596 pKa = 11.84SLGKK600 pKa = 9.08MCEE603 pKa = 4.12DD604 pKa = 3.88LVAGHH609 pKa = 6.07SEE611 pKa = 3.97EE612 pKa = 5.07QRR614 pKa = 11.84DD615 pKa = 3.77FLIKK619 pKa = 10.52RR620 pKa = 11.84FIQVNKK626 pKa = 9.27TLLAKK631 pKa = 10.31CPKK634 pKa = 8.8GQAWCIPRR642 pKa = 11.84RR643 pKa = 11.84LGGLGLPAPSGYY655 pKa = 9.96EE656 pKa = 3.67PRR658 pKa = 11.84PPQAKK663 pKa = 9.43LAAYY667 pKa = 9.08LATRR671 pKa = 11.84QLDD674 pKa = 3.84DD675 pKa = 4.71PDD677 pKa = 4.12LRR679 pKa = 11.84DD680 pKa = 3.59YY681 pKa = 11.12LYY683 pKa = 10.23PDD685 pKa = 3.09QPDD688 pKa = 3.73FLRR691 pKa = 11.84AYY693 pKa = 9.63ISYY696 pKa = 10.33RR697 pKa = 11.84QDD699 pKa = 2.72LCRR702 pKa = 11.84KK703 pKa = 9.98LGIEE707 pKa = 4.13STNEE711 pKa = 3.57QVVTLEE717 pKa = 4.84LPDD720 pKa = 3.79LALWAPMGAAGGDD733 pKa = 3.21KK734 pKa = 10.59TEE736 pKa = 4.03LVKK739 pKa = 10.89RR740 pKa = 11.84SIRR743 pKa = 11.84AWEE746 pKa = 4.02SLKK749 pKa = 10.83KK750 pKa = 10.36RR751 pKa = 11.84GLATSLHH758 pKa = 5.44AMKK761 pKa = 9.8MDD763 pKa = 5.04TIRR766 pKa = 11.84HH767 pKa = 4.41WTQEE771 pKa = 3.75PRR773 pKa = 11.84NIADD777 pKa = 3.36VSSWTVSWW785 pKa = 3.5
MM1 pKa = 7.86DD2 pKa = 6.2PIEE5 pKa = 4.43VSHH8 pKa = 6.95ALYY11 pKa = 10.52EE12 pKa = 4.88GLASWCMSRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 8.47GTKK26 pKa = 9.76DD27 pKa = 3.18PNFQDD32 pKa = 3.53LCRR35 pKa = 11.84LARR38 pKa = 11.84HH39 pKa = 6.51CGLCEE44 pKa = 3.79PPKK47 pKa = 10.8CLDD50 pKa = 3.34SALTEE55 pKa = 4.28FLNSFDD61 pKa = 4.25PPSCVEE67 pKa = 4.01HH68 pKa = 7.69DD69 pKa = 4.41NILKK73 pKa = 10.02EE74 pKa = 4.39RR75 pKa = 11.84IVHH78 pKa = 7.08SIQQANRR85 pKa = 11.84MVAVYY90 pKa = 10.27TDD92 pKa = 3.03ILKK95 pKa = 10.61FYY97 pKa = 10.36GVLGRR102 pKa = 11.84LHH104 pKa = 6.55PKK106 pKa = 8.68GWLWYY111 pKa = 9.74GVLGLLEE118 pKa = 5.75KK119 pKa = 10.25YY120 pKa = 10.32LKK122 pKa = 10.25WSTADD127 pKa = 3.45FLARR131 pKa = 11.84TLDD134 pKa = 4.33QEE136 pKa = 4.91DD137 pKa = 4.7LPPNPGFEE145 pKa = 4.47LGLPFTGPAASHH157 pKa = 6.83LKK159 pKa = 9.88RR160 pKa = 11.84VCLFNRR166 pKa = 11.84KK167 pKa = 9.18RR168 pKa = 11.84GLRR171 pKa = 11.84STYY174 pKa = 10.76AVALAYY180 pKa = 10.35SLYY183 pKa = 9.5QGKK186 pKa = 10.16GGSLPVSAEE195 pKa = 3.7FLEE198 pKa = 4.54NSVEE202 pKa = 4.06RR203 pKa = 11.84AVNALCTPRR212 pKa = 11.84TGMTEE217 pKa = 4.4DD218 pKa = 3.33IFDD221 pKa = 3.77VMSEE225 pKa = 3.96QVRR228 pKa = 11.84RR229 pKa = 11.84TVRR232 pKa = 11.84EE233 pKa = 3.91CFPRR237 pKa = 11.84HH238 pKa = 4.97HH239 pKa = 6.9SEE241 pKa = 3.69RR242 pKa = 11.84ARR244 pKa = 11.84PGSHH248 pKa = 7.41RR249 pKa = 11.84IPSLRR254 pKa = 11.84SSFQNSRR261 pKa = 11.84KK262 pKa = 9.44DD263 pKa = 3.04WGAFGHH269 pKa = 6.34IVSASISIGFPYY281 pKa = 10.93LLGHH285 pKa = 6.18WHH287 pKa = 6.89HH288 pKa = 6.46KK289 pKa = 8.86TNIEE293 pKa = 3.99LVYY296 pKa = 10.78GPSDD300 pKa = 3.81PEE302 pKa = 4.38DD303 pKa = 3.29VQLDD307 pKa = 3.84YY308 pKa = 10.98EE309 pKa = 4.2QSYY312 pKa = 10.19RR313 pKa = 11.84RR314 pKa = 11.84AVLMGRR320 pKa = 11.84CRR322 pKa = 11.84CYY324 pKa = 10.21PVGLLEE330 pKa = 4.16PFKK333 pKa = 11.21VRR335 pKa = 11.84VITRR339 pKa = 11.84GDD341 pKa = 3.03ADD343 pKa = 3.76MYY345 pKa = 10.73HH346 pKa = 7.53LARR349 pKa = 11.84RR350 pKa = 11.84WQKK353 pKa = 9.5TIHH356 pKa = 5.9GCLRR360 pKa = 11.84QHH362 pKa = 6.69PTFQLIGGPIDD373 pKa = 3.65WSALNHH379 pKa = 5.75LSEE382 pKa = 4.62RR383 pKa = 11.84TDD385 pKa = 3.13ICDD388 pKa = 3.21DD389 pKa = 4.26RR390 pKa = 11.84YY391 pKa = 10.03WVSADD396 pKa = 3.41YY397 pKa = 10.41TSATDD402 pKa = 3.75NLDD405 pKa = 3.75PEE407 pKa = 4.66LSRR410 pKa = 11.84VCMEE414 pKa = 5.24EE415 pKa = 3.11ICQRR419 pKa = 11.84LGVPLEE425 pKa = 4.37DD426 pKa = 3.34QLVLLRR432 pKa = 11.84TLTDD436 pKa = 3.13HH437 pKa = 7.68DD438 pKa = 5.11LYY440 pKa = 11.24EE441 pKa = 4.39EE442 pKa = 4.31SGDD445 pKa = 4.88LIGSQAWGQLMGSPSSFPILCLINAAATRR474 pKa = 11.84FALEE478 pKa = 3.7LANWKK483 pKa = 9.96VGEE486 pKa = 4.27YY487 pKa = 10.79KK488 pKa = 10.18LTDD491 pKa = 3.59LPMLFNGDD499 pKa = 3.55DD500 pKa = 3.32AAFRR504 pKa = 11.84ANEE507 pKa = 3.8QEE509 pKa = 3.88YY510 pKa = 8.91TIWKK514 pKa = 9.71GITKK518 pKa = 10.2SVGLEE523 pKa = 3.54FSVGKK528 pKa = 10.31NFFHH532 pKa = 7.47KK533 pKa = 10.45RR534 pKa = 11.84FLILNSAVHH543 pKa = 6.39EE544 pKa = 4.28DD545 pKa = 3.34VGRR548 pKa = 11.84VDD550 pKa = 3.62FFGRR554 pKa = 11.84RR555 pKa = 11.84FSRR558 pKa = 11.84LEE560 pKa = 3.7PVVTLSSSLLYY571 pKa = 10.65GQVRR575 pKa = 11.84VQSASQNDD583 pKa = 3.92RR584 pKa = 11.84THH586 pKa = 7.16LCQSDD591 pKa = 3.88LSVGRR596 pKa = 11.84SLGKK600 pKa = 9.08MCEE603 pKa = 4.12DD604 pKa = 3.88LVAGHH609 pKa = 6.07SEE611 pKa = 3.97EE612 pKa = 5.07QRR614 pKa = 11.84DD615 pKa = 3.77FLIKK619 pKa = 10.52RR620 pKa = 11.84FIQVNKK626 pKa = 9.27TLLAKK631 pKa = 10.31CPKK634 pKa = 8.8GQAWCIPRR642 pKa = 11.84RR643 pKa = 11.84LGGLGLPAPSGYY655 pKa = 9.96EE656 pKa = 3.67PRR658 pKa = 11.84PPQAKK663 pKa = 9.43LAAYY667 pKa = 9.08LATRR671 pKa = 11.84QLDD674 pKa = 3.84DD675 pKa = 4.71PDD677 pKa = 4.12LRR679 pKa = 11.84DD680 pKa = 3.59YY681 pKa = 11.12LYY683 pKa = 10.23PDD685 pKa = 3.09QPDD688 pKa = 3.73FLRR691 pKa = 11.84AYY693 pKa = 9.63ISYY696 pKa = 10.33RR697 pKa = 11.84QDD699 pKa = 2.72LCRR702 pKa = 11.84KK703 pKa = 9.98LGIEE707 pKa = 4.13STNEE711 pKa = 3.57QVVTLEE717 pKa = 4.84LPDD720 pKa = 3.79LALWAPMGAAGGDD733 pKa = 3.21KK734 pKa = 10.59TEE736 pKa = 4.03LVKK739 pKa = 10.89RR740 pKa = 11.84SIRR743 pKa = 11.84AWEE746 pKa = 4.02SLKK749 pKa = 10.83KK750 pKa = 10.36RR751 pKa = 11.84GLATSLHH758 pKa = 5.44AMKK761 pKa = 9.8MDD763 pKa = 5.04TIRR766 pKa = 11.84HH767 pKa = 4.41WTQEE771 pKa = 3.75PRR773 pKa = 11.84NIADD777 pKa = 3.36VSSWTVSWW785 pKa = 3.5
Molecular weight: 89.03 kDa
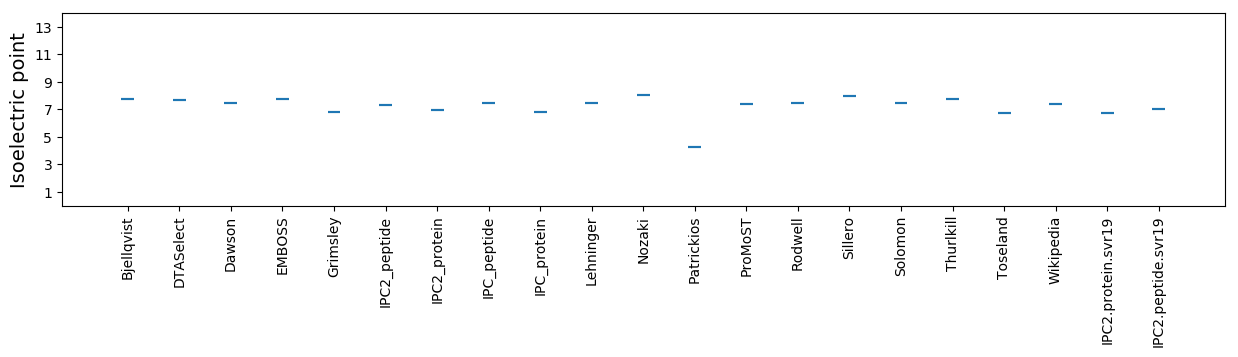
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI53|A0A1L3KI53_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 1 OX=1922436 PE=4 SV=1
MM1 pKa = 7.86DD2 pKa = 6.2PIEE5 pKa = 4.43VSHH8 pKa = 6.95ALYY11 pKa = 10.52EE12 pKa = 4.88GLASWCMSRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 8.47GTKK26 pKa = 9.76DD27 pKa = 3.18PNFQDD32 pKa = 3.53LCRR35 pKa = 11.84LARR38 pKa = 11.84HH39 pKa = 6.51CGLCEE44 pKa = 3.79PPKK47 pKa = 10.8CLDD50 pKa = 3.34SALTEE55 pKa = 4.28FLNSFDD61 pKa = 4.25PPSCVEE67 pKa = 4.01HH68 pKa = 7.69DD69 pKa = 4.41NILKK73 pKa = 10.02EE74 pKa = 4.39RR75 pKa = 11.84IVHH78 pKa = 7.08SIQQANRR85 pKa = 11.84MVAVYY90 pKa = 10.27TDD92 pKa = 3.03ILKK95 pKa = 10.61FYY97 pKa = 10.36GVLGRR102 pKa = 11.84LHH104 pKa = 6.55PKK106 pKa = 8.68GWLWYY111 pKa = 9.74GVLGLLEE118 pKa = 5.75KK119 pKa = 10.25YY120 pKa = 10.32LKK122 pKa = 10.25WSTADD127 pKa = 3.45FLARR131 pKa = 11.84TLDD134 pKa = 4.33QEE136 pKa = 4.91DD137 pKa = 4.7LPPNPGFEE145 pKa = 4.47LGLPFTGPAASHH157 pKa = 6.83LKK159 pKa = 9.88RR160 pKa = 11.84VCLFNRR166 pKa = 11.84KK167 pKa = 9.18RR168 pKa = 11.84GLRR171 pKa = 11.84STYY174 pKa = 10.76AVALAYY180 pKa = 10.35SLYY183 pKa = 9.5QGKK186 pKa = 10.16GGSLPVSAEE195 pKa = 3.7FLEE198 pKa = 4.54NSVEE202 pKa = 4.06RR203 pKa = 11.84AVNALCTPRR212 pKa = 11.84TGMTEE217 pKa = 4.4DD218 pKa = 3.33IFDD221 pKa = 3.77VMSEE225 pKa = 3.96QVRR228 pKa = 11.84RR229 pKa = 11.84TVRR232 pKa = 11.84EE233 pKa = 3.91CFPRR237 pKa = 11.84HH238 pKa = 4.97HH239 pKa = 6.9SEE241 pKa = 3.69RR242 pKa = 11.84ARR244 pKa = 11.84PGSHH248 pKa = 7.41RR249 pKa = 11.84IPSLRR254 pKa = 11.84SSFQNSRR261 pKa = 11.84KK262 pKa = 9.44DD263 pKa = 3.04WGAFGHH269 pKa = 6.34IVSASISIGFPYY281 pKa = 10.93LLGHH285 pKa = 6.18WHH287 pKa = 6.89HH288 pKa = 6.46KK289 pKa = 8.86TNIEE293 pKa = 3.99LVYY296 pKa = 10.78GPSDD300 pKa = 3.81PEE302 pKa = 4.38DD303 pKa = 3.29VQLDD307 pKa = 3.84YY308 pKa = 10.98EE309 pKa = 4.2QSYY312 pKa = 10.19RR313 pKa = 11.84RR314 pKa = 11.84AVLMGRR320 pKa = 11.84CRR322 pKa = 11.84CYY324 pKa = 10.21PVGLLEE330 pKa = 4.16PFKK333 pKa = 11.21VRR335 pKa = 11.84VITRR339 pKa = 11.84GDD341 pKa = 3.03ADD343 pKa = 3.76MYY345 pKa = 10.73HH346 pKa = 7.53LARR349 pKa = 11.84RR350 pKa = 11.84WQKK353 pKa = 9.5TIHH356 pKa = 5.9GCLRR360 pKa = 11.84QHH362 pKa = 6.69PTFQLIGGPIDD373 pKa = 3.65WSALNHH379 pKa = 5.75LSEE382 pKa = 4.62RR383 pKa = 11.84TDD385 pKa = 3.13ICDD388 pKa = 3.21DD389 pKa = 4.26RR390 pKa = 11.84YY391 pKa = 10.03WVSADD396 pKa = 3.41YY397 pKa = 10.41TSATDD402 pKa = 3.75NLDD405 pKa = 3.75PEE407 pKa = 4.66LSRR410 pKa = 11.84VCMEE414 pKa = 5.24EE415 pKa = 3.11ICQRR419 pKa = 11.84LGVPLEE425 pKa = 4.37DD426 pKa = 3.34QLVLLRR432 pKa = 11.84TLTDD436 pKa = 3.13HH437 pKa = 7.68DD438 pKa = 5.11LYY440 pKa = 11.24EE441 pKa = 4.39EE442 pKa = 4.31SGDD445 pKa = 4.88LIGSQAWGQLMGSPSSFPILCLINAAATRR474 pKa = 11.84FALEE478 pKa = 3.7LANWKK483 pKa = 9.96VGEE486 pKa = 4.27YY487 pKa = 10.79KK488 pKa = 10.18LTDD491 pKa = 3.59LPMLFNGDD499 pKa = 3.55DD500 pKa = 3.32AAFRR504 pKa = 11.84ANEE507 pKa = 3.8QEE509 pKa = 3.88YY510 pKa = 8.91TIWKK514 pKa = 9.71GITKK518 pKa = 10.2SVGLEE523 pKa = 3.54FSVGKK528 pKa = 10.31NFFHH532 pKa = 7.47KK533 pKa = 10.45RR534 pKa = 11.84FLILNSAVHH543 pKa = 6.39EE544 pKa = 4.28DD545 pKa = 3.34VGRR548 pKa = 11.84VDD550 pKa = 3.62FFGRR554 pKa = 11.84RR555 pKa = 11.84FSRR558 pKa = 11.84LEE560 pKa = 3.7PVVTLSSSLLYY571 pKa = 10.65GQVRR575 pKa = 11.84VQSASQNDD583 pKa = 3.92RR584 pKa = 11.84THH586 pKa = 7.16LCQSDD591 pKa = 3.88LSVGRR596 pKa = 11.84SLGKK600 pKa = 9.08MCEE603 pKa = 4.12DD604 pKa = 3.88LVAGHH609 pKa = 6.07SEE611 pKa = 3.97EE612 pKa = 5.07QRR614 pKa = 11.84DD615 pKa = 3.77FLIKK619 pKa = 10.52RR620 pKa = 11.84FIQVNKK626 pKa = 9.27TLLAKK631 pKa = 10.31CPKK634 pKa = 8.8GQAWCIPRR642 pKa = 11.84RR643 pKa = 11.84LGGLGLPAPSGYY655 pKa = 9.96EE656 pKa = 3.67PRR658 pKa = 11.84PPQAKK663 pKa = 9.43LAAYY667 pKa = 9.08LATRR671 pKa = 11.84QLDD674 pKa = 3.84DD675 pKa = 4.71PDD677 pKa = 4.12LRR679 pKa = 11.84DD680 pKa = 3.59YY681 pKa = 11.12LYY683 pKa = 10.23PDD685 pKa = 3.09QPDD688 pKa = 3.73FLRR691 pKa = 11.84AYY693 pKa = 9.63ISYY696 pKa = 10.33RR697 pKa = 11.84QDD699 pKa = 2.72LCRR702 pKa = 11.84KK703 pKa = 9.98LGIEE707 pKa = 4.13STNEE711 pKa = 3.57QVVTLEE717 pKa = 4.84LPDD720 pKa = 3.79LALWAPMGAAGGDD733 pKa = 3.21KK734 pKa = 10.59TEE736 pKa = 4.03LVKK739 pKa = 10.89RR740 pKa = 11.84SIRR743 pKa = 11.84AWEE746 pKa = 4.02SLKK749 pKa = 10.83KK750 pKa = 10.36RR751 pKa = 11.84GLATSLHH758 pKa = 5.44AMKK761 pKa = 9.8MDD763 pKa = 5.04TIRR766 pKa = 11.84HH767 pKa = 4.41WTQEE771 pKa = 3.75PRR773 pKa = 11.84NIADD777 pKa = 3.36VSSWTVSWW785 pKa = 3.5
MM1 pKa = 7.86DD2 pKa = 6.2PIEE5 pKa = 4.43VSHH8 pKa = 6.95ALYY11 pKa = 10.52EE12 pKa = 4.88GLASWCMSRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 8.47GTKK26 pKa = 9.76DD27 pKa = 3.18PNFQDD32 pKa = 3.53LCRR35 pKa = 11.84LARR38 pKa = 11.84HH39 pKa = 6.51CGLCEE44 pKa = 3.79PPKK47 pKa = 10.8CLDD50 pKa = 3.34SALTEE55 pKa = 4.28FLNSFDD61 pKa = 4.25PPSCVEE67 pKa = 4.01HH68 pKa = 7.69DD69 pKa = 4.41NILKK73 pKa = 10.02EE74 pKa = 4.39RR75 pKa = 11.84IVHH78 pKa = 7.08SIQQANRR85 pKa = 11.84MVAVYY90 pKa = 10.27TDD92 pKa = 3.03ILKK95 pKa = 10.61FYY97 pKa = 10.36GVLGRR102 pKa = 11.84LHH104 pKa = 6.55PKK106 pKa = 8.68GWLWYY111 pKa = 9.74GVLGLLEE118 pKa = 5.75KK119 pKa = 10.25YY120 pKa = 10.32LKK122 pKa = 10.25WSTADD127 pKa = 3.45FLARR131 pKa = 11.84TLDD134 pKa = 4.33QEE136 pKa = 4.91DD137 pKa = 4.7LPPNPGFEE145 pKa = 4.47LGLPFTGPAASHH157 pKa = 6.83LKK159 pKa = 9.88RR160 pKa = 11.84VCLFNRR166 pKa = 11.84KK167 pKa = 9.18RR168 pKa = 11.84GLRR171 pKa = 11.84STYY174 pKa = 10.76AVALAYY180 pKa = 10.35SLYY183 pKa = 9.5QGKK186 pKa = 10.16GGSLPVSAEE195 pKa = 3.7FLEE198 pKa = 4.54NSVEE202 pKa = 4.06RR203 pKa = 11.84AVNALCTPRR212 pKa = 11.84TGMTEE217 pKa = 4.4DD218 pKa = 3.33IFDD221 pKa = 3.77VMSEE225 pKa = 3.96QVRR228 pKa = 11.84RR229 pKa = 11.84TVRR232 pKa = 11.84EE233 pKa = 3.91CFPRR237 pKa = 11.84HH238 pKa = 4.97HH239 pKa = 6.9SEE241 pKa = 3.69RR242 pKa = 11.84ARR244 pKa = 11.84PGSHH248 pKa = 7.41RR249 pKa = 11.84IPSLRR254 pKa = 11.84SSFQNSRR261 pKa = 11.84KK262 pKa = 9.44DD263 pKa = 3.04WGAFGHH269 pKa = 6.34IVSASISIGFPYY281 pKa = 10.93LLGHH285 pKa = 6.18WHH287 pKa = 6.89HH288 pKa = 6.46KK289 pKa = 8.86TNIEE293 pKa = 3.99LVYY296 pKa = 10.78GPSDD300 pKa = 3.81PEE302 pKa = 4.38DD303 pKa = 3.29VQLDD307 pKa = 3.84YY308 pKa = 10.98EE309 pKa = 4.2QSYY312 pKa = 10.19RR313 pKa = 11.84RR314 pKa = 11.84AVLMGRR320 pKa = 11.84CRR322 pKa = 11.84CYY324 pKa = 10.21PVGLLEE330 pKa = 4.16PFKK333 pKa = 11.21VRR335 pKa = 11.84VITRR339 pKa = 11.84GDD341 pKa = 3.03ADD343 pKa = 3.76MYY345 pKa = 10.73HH346 pKa = 7.53LARR349 pKa = 11.84RR350 pKa = 11.84WQKK353 pKa = 9.5TIHH356 pKa = 5.9GCLRR360 pKa = 11.84QHH362 pKa = 6.69PTFQLIGGPIDD373 pKa = 3.65WSALNHH379 pKa = 5.75LSEE382 pKa = 4.62RR383 pKa = 11.84TDD385 pKa = 3.13ICDD388 pKa = 3.21DD389 pKa = 4.26RR390 pKa = 11.84YY391 pKa = 10.03WVSADD396 pKa = 3.41YY397 pKa = 10.41TSATDD402 pKa = 3.75NLDD405 pKa = 3.75PEE407 pKa = 4.66LSRR410 pKa = 11.84VCMEE414 pKa = 5.24EE415 pKa = 3.11ICQRR419 pKa = 11.84LGVPLEE425 pKa = 4.37DD426 pKa = 3.34QLVLLRR432 pKa = 11.84TLTDD436 pKa = 3.13HH437 pKa = 7.68DD438 pKa = 5.11LYY440 pKa = 11.24EE441 pKa = 4.39EE442 pKa = 4.31SGDD445 pKa = 4.88LIGSQAWGQLMGSPSSFPILCLINAAATRR474 pKa = 11.84FALEE478 pKa = 3.7LANWKK483 pKa = 9.96VGEE486 pKa = 4.27YY487 pKa = 10.79KK488 pKa = 10.18LTDD491 pKa = 3.59LPMLFNGDD499 pKa = 3.55DD500 pKa = 3.32AAFRR504 pKa = 11.84ANEE507 pKa = 3.8QEE509 pKa = 3.88YY510 pKa = 8.91TIWKK514 pKa = 9.71GITKK518 pKa = 10.2SVGLEE523 pKa = 3.54FSVGKK528 pKa = 10.31NFFHH532 pKa = 7.47KK533 pKa = 10.45RR534 pKa = 11.84FLILNSAVHH543 pKa = 6.39EE544 pKa = 4.28DD545 pKa = 3.34VGRR548 pKa = 11.84VDD550 pKa = 3.62FFGRR554 pKa = 11.84RR555 pKa = 11.84FSRR558 pKa = 11.84LEE560 pKa = 3.7PVVTLSSSLLYY571 pKa = 10.65GQVRR575 pKa = 11.84VQSASQNDD583 pKa = 3.92RR584 pKa = 11.84THH586 pKa = 7.16LCQSDD591 pKa = 3.88LSVGRR596 pKa = 11.84SLGKK600 pKa = 9.08MCEE603 pKa = 4.12DD604 pKa = 3.88LVAGHH609 pKa = 6.07SEE611 pKa = 3.97EE612 pKa = 5.07QRR614 pKa = 11.84DD615 pKa = 3.77FLIKK619 pKa = 10.52RR620 pKa = 11.84FIQVNKK626 pKa = 9.27TLLAKK631 pKa = 10.31CPKK634 pKa = 8.8GQAWCIPRR642 pKa = 11.84RR643 pKa = 11.84LGGLGLPAPSGYY655 pKa = 9.96EE656 pKa = 3.67PRR658 pKa = 11.84PPQAKK663 pKa = 9.43LAAYY667 pKa = 9.08LATRR671 pKa = 11.84QLDD674 pKa = 3.84DD675 pKa = 4.71PDD677 pKa = 4.12LRR679 pKa = 11.84DD680 pKa = 3.59YY681 pKa = 11.12LYY683 pKa = 10.23PDD685 pKa = 3.09QPDD688 pKa = 3.73FLRR691 pKa = 11.84AYY693 pKa = 9.63ISYY696 pKa = 10.33RR697 pKa = 11.84QDD699 pKa = 2.72LCRR702 pKa = 11.84KK703 pKa = 9.98LGIEE707 pKa = 4.13STNEE711 pKa = 3.57QVVTLEE717 pKa = 4.84LPDD720 pKa = 3.79LALWAPMGAAGGDD733 pKa = 3.21KK734 pKa = 10.59TEE736 pKa = 4.03LVKK739 pKa = 10.89RR740 pKa = 11.84SIRR743 pKa = 11.84AWEE746 pKa = 4.02SLKK749 pKa = 10.83KK750 pKa = 10.36RR751 pKa = 11.84GLATSLHH758 pKa = 5.44AMKK761 pKa = 9.8MDD763 pKa = 5.04TIRR766 pKa = 11.84HH767 pKa = 4.41WTQEE771 pKa = 3.75PRR773 pKa = 11.84NIADD777 pKa = 3.36VSSWTVSWW785 pKa = 3.5
Molecular weight: 89.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
785 |
785 |
785 |
785.0 |
89.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.752 ± 0.0 | 2.675 ± 0.0 |
6.242 ± 0.0 | 5.605 ± 0.0 |
3.822 ± 0.0 | 6.879 ± 0.0 |
3.057 ± 0.0 | 3.949 ± 0.0 |
4.204 ± 0.0 | 11.975 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.783 ± 0.0 | 2.803 ± 0.0 |
5.478 ± 0.0 | 3.822 ± 0.0 |
7.898 ± 0.0 | 7.389 ± 0.0 |
4.459 ± 0.0 | 5.605 ± 0.0 |
2.293 ± 0.0 | 3.312 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
