
Capybara microvirus Cap3_SP_554
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.45
Get precalculated fractions of proteins
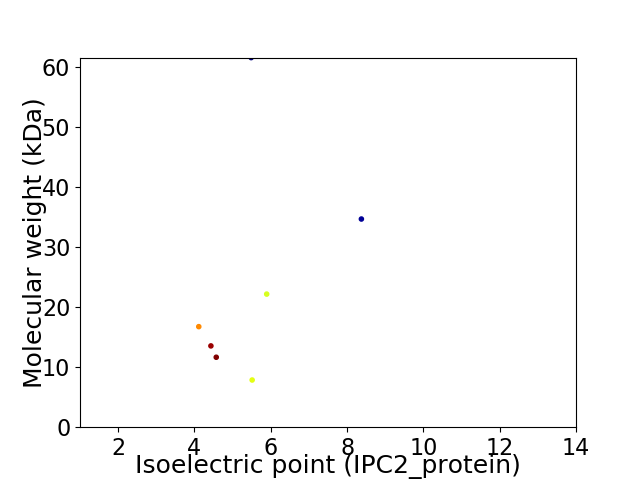
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W656|A0A4P8W656_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_554 OX=2585475 PE=4 SV=1
MM1 pKa = 7.74AFLKK5 pKa = 10.38KK6 pKa = 10.1QYY8 pKa = 8.19EE9 pKa = 4.35VKK11 pKa = 10.88YY12 pKa = 8.18PTFSNIVPDD21 pKa = 3.54YY22 pKa = 10.45KK23 pKa = 10.97YY24 pKa = 11.25NEE26 pKa = 3.83VTGIVEE32 pKa = 4.31EE33 pKa = 4.21VGKK36 pKa = 10.33IDD38 pKa = 3.94LQEE41 pKa = 4.87KK42 pKa = 10.25INSCRR47 pKa = 11.84VNCLDD52 pKa = 5.04YY53 pKa = 11.16ILDD56 pKa = 3.65QYY58 pKa = 11.38LEE60 pKa = 4.49NGNSVNDD67 pKa = 3.87LFKK70 pKa = 11.32VSVDD74 pKa = 3.53VSDD77 pKa = 6.24DD78 pKa = 3.45IADD81 pKa = 4.09CSQPLDD87 pKa = 4.56DD88 pKa = 5.99IMYY91 pKa = 10.6QNDD94 pKa = 3.32VMNLMDD100 pKa = 4.58EE101 pKa = 5.19LKK103 pKa = 10.54DD104 pKa = 3.52KK105 pKa = 11.45YY106 pKa = 10.76GASDD110 pKa = 3.42MSNDD114 pKa = 4.04DD115 pKa = 3.92FIKK118 pKa = 10.77YY119 pKa = 10.58LSDD122 pKa = 3.3EE123 pKa = 4.49KK124 pKa = 11.02IKK126 pKa = 10.42IDD128 pKa = 3.39TYY130 pKa = 11.09INEE133 pKa = 4.03KK134 pKa = 8.25THH136 pKa = 6.74AKK138 pKa = 10.12EE139 pKa = 3.68GDD141 pKa = 3.5INEE144 pKa = 4.34SS145 pKa = 3.23
MM1 pKa = 7.74AFLKK5 pKa = 10.38KK6 pKa = 10.1QYY8 pKa = 8.19EE9 pKa = 4.35VKK11 pKa = 10.88YY12 pKa = 8.18PTFSNIVPDD21 pKa = 3.54YY22 pKa = 10.45KK23 pKa = 10.97YY24 pKa = 11.25NEE26 pKa = 3.83VTGIVEE32 pKa = 4.31EE33 pKa = 4.21VGKK36 pKa = 10.33IDD38 pKa = 3.94LQEE41 pKa = 4.87KK42 pKa = 10.25INSCRR47 pKa = 11.84VNCLDD52 pKa = 5.04YY53 pKa = 11.16ILDD56 pKa = 3.65QYY58 pKa = 11.38LEE60 pKa = 4.49NGNSVNDD67 pKa = 3.87LFKK70 pKa = 11.32VSVDD74 pKa = 3.53VSDD77 pKa = 6.24DD78 pKa = 3.45IADD81 pKa = 4.09CSQPLDD87 pKa = 4.56DD88 pKa = 5.99IMYY91 pKa = 10.6QNDD94 pKa = 3.32VMNLMDD100 pKa = 4.58EE101 pKa = 5.19LKK103 pKa = 10.54DD104 pKa = 3.52KK105 pKa = 11.45YY106 pKa = 10.76GASDD110 pKa = 3.42MSNDD114 pKa = 4.04DD115 pKa = 3.92FIKK118 pKa = 10.77YY119 pKa = 10.58LSDD122 pKa = 3.3EE123 pKa = 4.49KK124 pKa = 11.02IKK126 pKa = 10.42IDD128 pKa = 3.39TYY130 pKa = 11.09INEE133 pKa = 4.03KK134 pKa = 8.25THH136 pKa = 6.74AKK138 pKa = 10.12EE139 pKa = 3.68GDD141 pKa = 3.5INEE144 pKa = 4.34SS145 pKa = 3.23
Molecular weight: 16.74 kDa
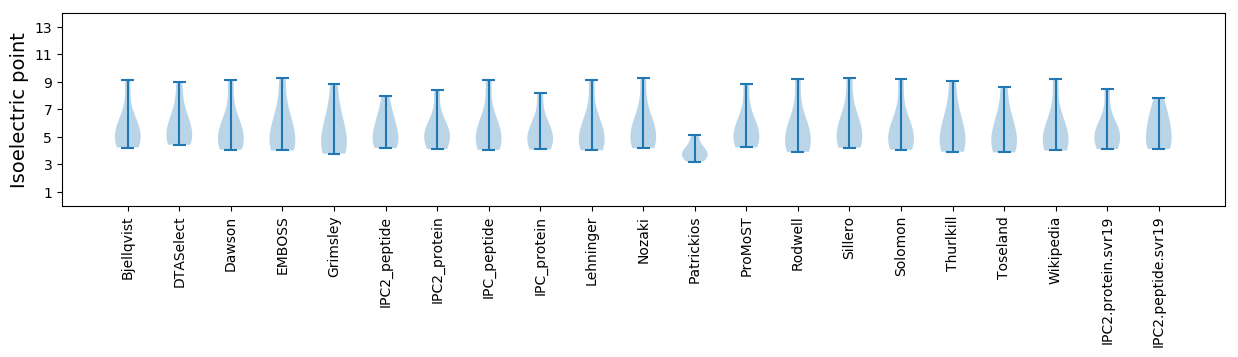
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVU7|A0A4V1FVU7_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_554 OX=2585475 PE=4 SV=1
MM1 pKa = 7.53CLSPLKK7 pKa = 10.25AVHH10 pKa = 6.63SVFEE14 pKa = 4.36DD15 pKa = 3.99LEE17 pKa = 4.41TGEE20 pKa = 4.23VHH22 pKa = 7.36SKK24 pKa = 10.55IKK26 pKa = 8.13ITKK29 pKa = 9.83YY30 pKa = 8.92PDD32 pKa = 2.97LFIRR36 pKa = 11.84DD37 pKa = 3.59NPTIIKK43 pKa = 9.8CEE45 pKa = 4.61PISLPCGHH53 pKa = 7.22CIEE56 pKa = 5.16CLNQISNEE64 pKa = 3.68WTFRR68 pKa = 11.84LQAEE72 pKa = 4.6SIYY75 pKa = 10.41HH76 pKa = 5.9SKK78 pKa = 11.06ACFITLTYY86 pKa = 11.0AEE88 pKa = 4.81TDD90 pKa = 3.15GSLNPRR96 pKa = 11.84DD97 pKa = 3.27IEE99 pKa = 4.32LFLKK103 pKa = 10.55RR104 pKa = 11.84LRR106 pKa = 11.84KK107 pKa = 10.02SIAPVKK113 pKa = 10.02IRR115 pKa = 11.84YY116 pKa = 7.97FLSGEE121 pKa = 4.13YY122 pKa = 10.12GSKK125 pKa = 10.33GLRR128 pKa = 11.84PHH130 pKa = 5.34YY131 pKa = 10.16HH132 pKa = 5.76VIVFGWCPDD141 pKa = 3.79DD142 pKa = 4.58LYY144 pKa = 11.29FWKK147 pKa = 10.32KK148 pKa = 9.32SKK150 pKa = 10.56KK151 pKa = 10.21GFINYY156 pKa = 8.38RR157 pKa = 11.84SPKK160 pKa = 9.7VEE162 pKa = 4.2KK163 pKa = 9.62LWKK166 pKa = 10.03FGFSTVEE173 pKa = 3.86MLSSRR178 pKa = 11.84TCKK181 pKa = 9.48YY182 pKa = 6.96TAKK185 pKa = 10.35YY186 pKa = 6.83MQKK189 pKa = 7.9MTLNWEE195 pKa = 4.32TNHH198 pKa = 5.68VYY200 pKa = 10.5PFVRR204 pKa = 11.84MSTKK208 pKa = 10.27PGIGFQWFLDD218 pKa = 3.51HH219 pKa = 7.06KK220 pKa = 10.58RR221 pKa = 11.84CLEE224 pKa = 3.97TDD226 pKa = 2.99TMYY229 pKa = 10.62IGGIGRR235 pKa = 11.84RR236 pKa = 11.84VPRR239 pKa = 11.84YY240 pKa = 8.31FLKK243 pKa = 9.72IAEE246 pKa = 4.18RR247 pKa = 11.84EE248 pKa = 4.27GVNLSEE254 pKa = 4.8LKK256 pKa = 10.57EE257 pKa = 3.97NRR259 pKa = 11.84INLAIKK265 pKa = 9.55FEE267 pKa = 4.43RR268 pKa = 11.84DD269 pKa = 3.31SITLEE274 pKa = 3.87NLKK277 pKa = 10.58NKK279 pKa = 10.28YY280 pKa = 10.19LDD282 pKa = 5.52LFGAWCYY289 pKa = 10.91NDD291 pKa = 3.75SGGEE295 pKa = 4.0KK296 pKa = 10.36
MM1 pKa = 7.53CLSPLKK7 pKa = 10.25AVHH10 pKa = 6.63SVFEE14 pKa = 4.36DD15 pKa = 3.99LEE17 pKa = 4.41TGEE20 pKa = 4.23VHH22 pKa = 7.36SKK24 pKa = 10.55IKK26 pKa = 8.13ITKK29 pKa = 9.83YY30 pKa = 8.92PDD32 pKa = 2.97LFIRR36 pKa = 11.84DD37 pKa = 3.59NPTIIKK43 pKa = 9.8CEE45 pKa = 4.61PISLPCGHH53 pKa = 7.22CIEE56 pKa = 5.16CLNQISNEE64 pKa = 3.68WTFRR68 pKa = 11.84LQAEE72 pKa = 4.6SIYY75 pKa = 10.41HH76 pKa = 5.9SKK78 pKa = 11.06ACFITLTYY86 pKa = 11.0AEE88 pKa = 4.81TDD90 pKa = 3.15GSLNPRR96 pKa = 11.84DD97 pKa = 3.27IEE99 pKa = 4.32LFLKK103 pKa = 10.55RR104 pKa = 11.84LRR106 pKa = 11.84KK107 pKa = 10.02SIAPVKK113 pKa = 10.02IRR115 pKa = 11.84YY116 pKa = 7.97FLSGEE121 pKa = 4.13YY122 pKa = 10.12GSKK125 pKa = 10.33GLRR128 pKa = 11.84PHH130 pKa = 5.34YY131 pKa = 10.16HH132 pKa = 5.76VIVFGWCPDD141 pKa = 3.79DD142 pKa = 4.58LYY144 pKa = 11.29FWKK147 pKa = 10.32KK148 pKa = 9.32SKK150 pKa = 10.56KK151 pKa = 10.21GFINYY156 pKa = 8.38RR157 pKa = 11.84SPKK160 pKa = 9.7VEE162 pKa = 4.2KK163 pKa = 9.62LWKK166 pKa = 10.03FGFSTVEE173 pKa = 3.86MLSSRR178 pKa = 11.84TCKK181 pKa = 9.48YY182 pKa = 6.96TAKK185 pKa = 10.35YY186 pKa = 6.83MQKK189 pKa = 7.9MTLNWEE195 pKa = 4.32TNHH198 pKa = 5.68VYY200 pKa = 10.5PFVRR204 pKa = 11.84MSTKK208 pKa = 10.27PGIGFQWFLDD218 pKa = 3.51HH219 pKa = 7.06KK220 pKa = 10.58RR221 pKa = 11.84CLEE224 pKa = 3.97TDD226 pKa = 2.99TMYY229 pKa = 10.62IGGIGRR235 pKa = 11.84RR236 pKa = 11.84VPRR239 pKa = 11.84YY240 pKa = 8.31FLKK243 pKa = 9.72IAEE246 pKa = 4.18RR247 pKa = 11.84EE248 pKa = 4.27GVNLSEE254 pKa = 4.8LKK256 pKa = 10.57EE257 pKa = 3.97NRR259 pKa = 11.84INLAIKK265 pKa = 9.55FEE267 pKa = 4.43RR268 pKa = 11.84DD269 pKa = 3.31SITLEE274 pKa = 3.87NLKK277 pKa = 10.58NKK279 pKa = 10.28YY280 pKa = 10.19LDD282 pKa = 5.52LFGAWCYY289 pKa = 10.91NDD291 pKa = 3.75SGGEE295 pKa = 4.0KK296 pKa = 10.36
Molecular weight: 34.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1490 |
71 |
553 |
212.9 |
24.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.906 ± 1.427 | 1.477 ± 0.616 |
7.047 ± 1.034 | 5.302 ± 0.879 |
5.235 ± 0.521 | 6.107 ± 0.73 |
1.678 ± 0.352 | 8.255 ± 0.694 |
6.846 ± 1.171 | 7.718 ± 0.927 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.349 ± 0.398 | 7.181 ± 1.007 |
3.758 ± 0.761 | 2.416 ± 0.298 |
3.557 ± 0.656 | 8.658 ± 0.82 |
4.43 ± 0.598 | 5.973 ± 0.777 |
1.074 ± 0.311 | 5.034 ± 0.331 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
