
Bifiguratus adelaidae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Mucoromycota; Mucoromycotina; Endogonomycetes; Endogonales; Endogonales incertae sedis; Bifiguratus
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
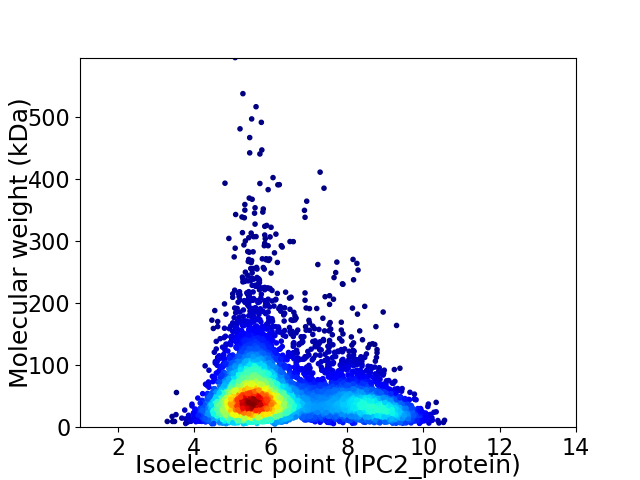
Virtual 2D-PAGE plot for 5718 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A261XVQ4|A0A261XVQ4_9FUNG V-type proton ATPase subunit a OS=Bifiguratus adelaidae OX=1938954 GN=BZG36_04462 PE=3 SV=1
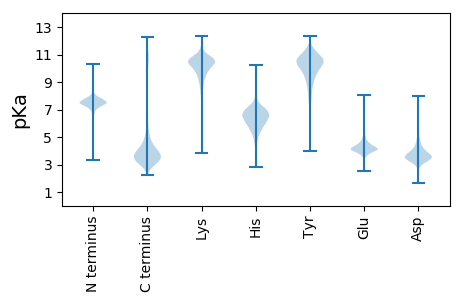
MM1 pKa = 7.52NFDD4 pKa = 3.5QQQQQGSLGQRR15 pKa = 11.84QSGLNRR21 pKa = 11.84QEE23 pKa = 3.99EE24 pKa = 5.74GYY26 pKa = 10.07GQSDD30 pKa = 3.99YY31 pKa = 11.29GQQGDD36 pKa = 3.78ISQQNMGSDD45 pKa = 3.41SGQFGGMQPSYY56 pKa = 10.98GGNTQQPYY64 pKa = 10.33SSSDD68 pKa = 3.33YY69 pKa = 10.62SQQDD73 pKa = 3.19PSQQLGSQGQQQGQKK88 pKa = 9.11SQGGEE93 pKa = 3.96SQMQAPSAGSDD104 pKa = 2.94NTQFIDD110 pKa = 4.08ASQRR114 pKa = 11.84DD115 pKa = 3.82QQQGSNQNWW124 pKa = 3.1
MM1 pKa = 7.52NFDD4 pKa = 3.5QQQQQGSLGQRR15 pKa = 11.84QSGLNRR21 pKa = 11.84QEE23 pKa = 3.99EE24 pKa = 5.74GYY26 pKa = 10.07GQSDD30 pKa = 3.99YY31 pKa = 11.29GQQGDD36 pKa = 3.78ISQQNMGSDD45 pKa = 3.41SGQFGGMQPSYY56 pKa = 10.98GGNTQQPYY64 pKa = 10.33SSSDD68 pKa = 3.33YY69 pKa = 10.62SQQDD73 pKa = 3.19PSQQLGSQGQQQGQKK88 pKa = 9.11SQGGEE93 pKa = 3.96SQMQAPSAGSDD104 pKa = 2.94NTQFIDD110 pKa = 4.08ASQRR114 pKa = 11.84DD115 pKa = 3.82QQQGSNQNWW124 pKa = 3.1
Molecular weight: 13.45 kDa
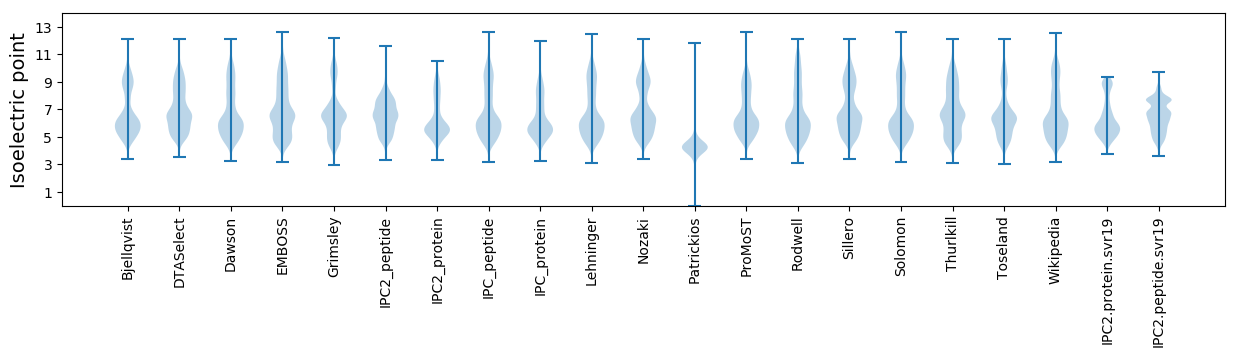
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A261XZT6|A0A261XZT6_9FUNG Uncharacterized protein OS=Bifiguratus adelaidae OX=1938954 GN=BZG36_03679 PE=4 SV=1
MM1 pKa = 6.95STPRR5 pKa = 11.84LTSFRR10 pKa = 11.84RR11 pKa = 11.84FAVISGHH18 pKa = 6.46PPFAAQSPLAAGQVRR33 pKa = 11.84LLHH36 pKa = 6.45RR37 pKa = 11.84SSNRR41 pKa = 11.84YY42 pKa = 7.98FKK44 pKa = 10.7CQSAVAQTPRR54 pKa = 11.84AEE56 pKa = 4.22PGIIATIQRR65 pKa = 11.84DD66 pKa = 3.9FKK68 pKa = 11.3SIFPRR73 pKa = 11.84RR74 pKa = 11.84SSPLGHH80 pKa = 6.16RR81 pKa = 11.84RR82 pKa = 11.84HH83 pKa = 6.17RR84 pKa = 11.84KK85 pKa = 8.91GRR87 pKa = 11.84ALWAAVLQCRR97 pKa = 11.84KK98 pKa = 8.86WLNDD102 pKa = 3.21PFSIQFRR109 pKa = 11.84TPRR112 pKa = 11.84LFQVVAATYY121 pKa = 9.32QQFLRR126 pKa = 11.84DD127 pKa = 3.3IYY129 pKa = 10.63HH130 pKa = 6.58IYY132 pKa = 10.43DD133 pKa = 3.69RR134 pKa = 11.84PSTVDD139 pKa = 2.92MNYY142 pKa = 10.98AMSAKK147 pKa = 10.23SLFWNSRR154 pKa = 11.84WNHH157 pKa = 4.96RR158 pKa = 11.84LKK160 pKa = 11.11VPLAGPILDD169 pKa = 3.75AMTPEE174 pKa = 4.52SLHH177 pKa = 5.85TMIALPFATPKK188 pKa = 10.6HH189 pKa = 5.68LVFEE193 pKa = 4.44YY194 pKa = 11.03GDD196 pKa = 3.74MRR198 pKa = 11.84IHH200 pKa = 7.88ASRR203 pKa = 11.84NSLLSIADD211 pKa = 4.22TITSKK216 pKa = 11.1GSTSAFILTNINILKK231 pKa = 10.19ADD233 pKa = 4.33LDD235 pKa = 4.97AICSSPSSQRR245 pKa = 11.84IKK247 pKa = 10.66RR248 pKa = 11.84LPNAGGSSHH257 pKa = 6.57KK258 pKa = 10.82SEE260 pKa = 3.92VLSYY264 pKa = 11.22VFLEE268 pKa = 4.12RR269 pKa = 11.84MLGCEE274 pKa = 3.98LLKK277 pKa = 10.36TEE279 pKa = 4.53MEE281 pKa = 4.57IIYY284 pKa = 9.26EE285 pKa = 3.98PRR287 pKa = 11.84GGPMVDD293 pKa = 3.64YY294 pKa = 10.67SIHH297 pKa = 6.53LPQHH301 pKa = 6.68LGSTWLAVSVTRR313 pKa = 11.84AIAFGRR319 pKa = 11.84NFTLQDD325 pKa = 3.14AEE327 pKa = 4.02KK328 pKa = 10.44LIRR331 pKa = 11.84HH332 pKa = 6.19KK333 pKa = 11.02LLGVFFSNQTIVQRR347 pKa = 11.84QRR349 pKa = 11.84PSRR352 pKa = 11.84QILHH356 pKa = 5.04IWARR360 pKa = 11.84EE361 pKa = 3.91GKK363 pKa = 8.97VARR366 pKa = 11.84ILSKK370 pKa = 10.13AWSRR374 pKa = 11.84VMDD377 pKa = 5.05DD378 pKa = 3.95MPKK381 pKa = 8.23STIVIVSVVNMDD393 pKa = 3.87AVFLNKK399 pKa = 10.0AFKK402 pKa = 10.2QQ403 pKa = 3.62
MM1 pKa = 6.95STPRR5 pKa = 11.84LTSFRR10 pKa = 11.84RR11 pKa = 11.84FAVISGHH18 pKa = 6.46PPFAAQSPLAAGQVRR33 pKa = 11.84LLHH36 pKa = 6.45RR37 pKa = 11.84SSNRR41 pKa = 11.84YY42 pKa = 7.98FKK44 pKa = 10.7CQSAVAQTPRR54 pKa = 11.84AEE56 pKa = 4.22PGIIATIQRR65 pKa = 11.84DD66 pKa = 3.9FKK68 pKa = 11.3SIFPRR73 pKa = 11.84RR74 pKa = 11.84SSPLGHH80 pKa = 6.16RR81 pKa = 11.84RR82 pKa = 11.84HH83 pKa = 6.17RR84 pKa = 11.84KK85 pKa = 8.91GRR87 pKa = 11.84ALWAAVLQCRR97 pKa = 11.84KK98 pKa = 8.86WLNDD102 pKa = 3.21PFSIQFRR109 pKa = 11.84TPRR112 pKa = 11.84LFQVVAATYY121 pKa = 9.32QQFLRR126 pKa = 11.84DD127 pKa = 3.3IYY129 pKa = 10.63HH130 pKa = 6.58IYY132 pKa = 10.43DD133 pKa = 3.69RR134 pKa = 11.84PSTVDD139 pKa = 2.92MNYY142 pKa = 10.98AMSAKK147 pKa = 10.23SLFWNSRR154 pKa = 11.84WNHH157 pKa = 4.96RR158 pKa = 11.84LKK160 pKa = 11.11VPLAGPILDD169 pKa = 3.75AMTPEE174 pKa = 4.52SLHH177 pKa = 5.85TMIALPFATPKK188 pKa = 10.6HH189 pKa = 5.68LVFEE193 pKa = 4.44YY194 pKa = 11.03GDD196 pKa = 3.74MRR198 pKa = 11.84IHH200 pKa = 7.88ASRR203 pKa = 11.84NSLLSIADD211 pKa = 4.22TITSKK216 pKa = 11.1GSTSAFILTNINILKK231 pKa = 10.19ADD233 pKa = 4.33LDD235 pKa = 4.97AICSSPSSQRR245 pKa = 11.84IKK247 pKa = 10.66RR248 pKa = 11.84LPNAGGSSHH257 pKa = 6.57KK258 pKa = 10.82SEE260 pKa = 3.92VLSYY264 pKa = 11.22VFLEE268 pKa = 4.12RR269 pKa = 11.84MLGCEE274 pKa = 3.98LLKK277 pKa = 10.36TEE279 pKa = 4.53MEE281 pKa = 4.57IIYY284 pKa = 9.26EE285 pKa = 3.98PRR287 pKa = 11.84GGPMVDD293 pKa = 3.64YY294 pKa = 10.67SIHH297 pKa = 6.53LPQHH301 pKa = 6.68LGSTWLAVSVTRR313 pKa = 11.84AIAFGRR319 pKa = 11.84NFTLQDD325 pKa = 3.14AEE327 pKa = 4.02KK328 pKa = 10.44LIRR331 pKa = 11.84HH332 pKa = 6.19KK333 pKa = 11.02LLGVFFSNQTIVQRR347 pKa = 11.84QRR349 pKa = 11.84PSRR352 pKa = 11.84QILHH356 pKa = 5.04IWARR360 pKa = 11.84EE361 pKa = 3.91GKK363 pKa = 8.97VARR366 pKa = 11.84ILSKK370 pKa = 10.13AWSRR374 pKa = 11.84VMDD377 pKa = 5.05DD378 pKa = 3.95MPKK381 pKa = 8.23STIVIVSVVNMDD393 pKa = 3.87AVFLNKK399 pKa = 10.0AFKK402 pKa = 10.2QQ403 pKa = 3.62
Molecular weight: 45.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3128832 |
51 |
5291 |
547.2 |
61.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.006 ± 0.022 | 1.222 ± 0.01 |
5.9 ± 0.018 | 6.19 ± 0.028 |
3.788 ± 0.017 | 6.064 ± 0.028 |
2.575 ± 0.013 | 5.301 ± 0.019 |
5.263 ± 0.022 | 9.135 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.357 ± 0.012 | 4.039 ± 0.018 |
5.437 ± 0.027 | 4.369 ± 0.017 |
5.638 ± 0.023 | 7.947 ± 0.029 |
6.069 ± 0.016 | 6.289 ± 0.016 |
1.272 ± 0.01 | 3.138 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
