
Lutibacter sp. Hel_I_33_5
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Lutibacter; unclassified Lutibacter
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
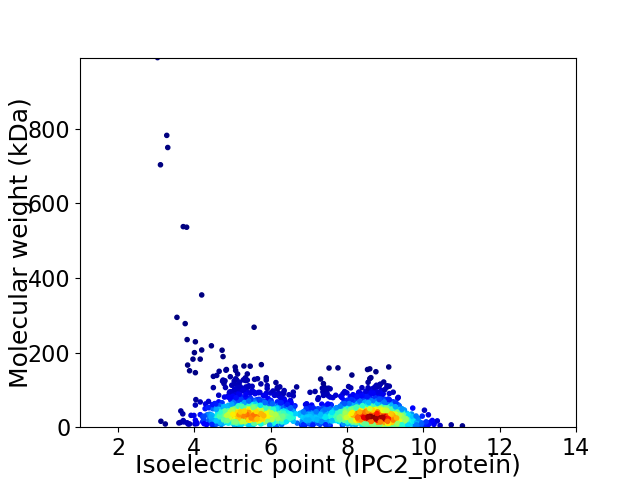
Virtual 2D-PAGE plot for 2634 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A559RYQ8|A0A559RYQ8_9FLAO Uncharacterized protein OS=Lutibacter sp. Hel_I_33_5 OX=1566289 GN=OD91_0488 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.52NNYY5 pKa = 9.87SKK7 pKa = 11.13FILKK11 pKa = 8.86TLLLFAILFGLEE23 pKa = 3.88TYY25 pKa = 10.01AQQNIIAQTQNITVQLDD42 pKa = 3.56ANGEE46 pKa = 4.25AIITPQQINNGSTATTGIQSLNLDD70 pKa = 3.62TSDD73 pKa = 5.29FNCTDD78 pKa = 2.94INGDD82 pKa = 3.69RR83 pKa = 11.84NFSYY87 pKa = 11.0LLDD90 pKa = 4.5GEE92 pKa = 4.49TDD94 pKa = 3.23ATGDD98 pKa = 4.04EE99 pKa = 4.34ILVDD103 pKa = 3.93DD104 pKa = 4.79TNLTTLTTQTIEE116 pKa = 3.44AWIRR120 pKa = 11.84PATGASDD127 pKa = 4.0NDD129 pKa = 3.31VLVAVRR135 pKa = 11.84NNIHH139 pKa = 6.24NSRR142 pKa = 11.84FSIHH146 pKa = 6.62INADD150 pKa = 3.29TNTIGIFNSTNWQTITTIINANQWYY175 pKa = 9.43HH176 pKa = 6.08IALVMNSTKK185 pKa = 10.36TLVYY189 pKa = 10.51LDD191 pKa = 3.92GAFEE195 pKa = 4.68GEE197 pKa = 3.9ISTGINTSLGANAPLSIGSPNDD219 pKa = 3.44TGWYY223 pKa = 7.61SEE225 pKa = 3.87KK226 pKa = 10.41WEE228 pKa = 4.36GNIDD232 pKa = 5.46DD233 pKa = 4.21LRR235 pKa = 11.84IWNDD239 pKa = 2.44EE240 pKa = 3.95RR241 pKa = 11.84TALEE245 pKa = 3.64IANNRR250 pKa = 11.84NIEE253 pKa = 4.11LVGNEE258 pKa = 4.22AGLVGYY264 pKa = 10.48FDD266 pKa = 5.05FNEE269 pKa = 4.58GTGNTITNLAGTVNGTANNSQNANWSTDD297 pKa = 3.14SPIEE301 pKa = 4.42SNGNTVNLTVTDD313 pKa = 3.86VNGNSATTPANVIVVDD329 pKa = 5.01NIAPIAPTLADD340 pKa = 3.43VTGEE344 pKa = 4.02CSATVTAPTTTDD356 pKa = 2.77NCAGTITGTTTDD368 pKa = 3.32SLTYY372 pKa = 7.42TTQGTHH378 pKa = 6.68IITWNFDD385 pKa = 3.58DD386 pKa = 5.63GNGNSTTASQNVIISDD402 pKa = 3.98STPPTAITNNISVNLDD418 pKa = 3.23ANGNASITPQQIDD431 pKa = 3.55NGSSDD436 pKa = 3.43ACGISSISCSRR447 pKa = 11.84VGGSGNGLTNFLNGTLNNGTADD469 pKa = 3.21IIEE472 pKa = 4.33YY473 pKa = 10.71SNSNAPNYY481 pKa = 7.41RR482 pKa = 11.84TSFGTTCNEE491 pKa = 3.02AWFARR496 pKa = 11.84GMLCDD501 pKa = 4.66LAPNQNDD508 pKa = 3.73PDD510 pKa = 3.71NSSRR514 pKa = 11.84QDD516 pKa = 3.41NNAGATWRR524 pKa = 11.84NTSPGNSYY532 pKa = 10.56GRR534 pKa = 11.84LVIDD538 pKa = 4.97LGSSQTINAMSIFQMFSDD556 pKa = 4.19GKK558 pKa = 6.81TTHH561 pKa = 6.72IEE563 pKa = 3.62AFAYY567 pKa = 10.21NGNLEE572 pKa = 4.36NPNSSSPGWNQLFPSTSVGAGVLNGTTVSSPLKK605 pKa = 8.72TTFQSTNTRR614 pKa = 11.84FIKK617 pKa = 10.58LYY619 pKa = 10.44AKK621 pKa = 10.52NDD623 pKa = 3.28GSLGFSGYY631 pKa = 9.92IEE633 pKa = 4.04LRR635 pKa = 11.84QVKK638 pKa = 8.98VFNTNEE644 pKa = 3.77SNACDD649 pKa = 4.49FNCDD653 pKa = 3.79DD654 pKa = 4.63LGEE657 pKa = 4.41NNVTLTVEE665 pKa = 4.49DD666 pKa = 3.87KK667 pKa = 11.19NGNISTKK674 pKa = 10.5DD675 pKa = 3.51AIVTIIDD682 pKa = 4.82AIAPALEE689 pKa = 4.32ATPADD694 pKa = 3.89LTVEE698 pKa = 4.25CHH700 pKa = 6.73SIPDD704 pKa = 3.56VATISATDD712 pKa = 3.38NCDD715 pKa = 3.33SNISVNFTEE724 pKa = 4.09QRR726 pKa = 11.84ADD728 pKa = 3.23GFSNDD733 pKa = 3.57NYY735 pKa = 10.28TLTRR739 pKa = 11.84TWSATDD745 pKa = 3.21QSGNTVSEE753 pKa = 4.39SQVITVQDD761 pKa = 3.69VTPPTIVGQNISVTLTASGTVSIIPQNVLNNGSDD795 pKa = 3.37TCSTVSYY802 pKa = 9.46TISQNTFGATDD813 pKa = 5.5AINSPVTIQLIGTDD827 pKa = 3.51EE828 pKa = 4.43SGNATTIPVQVTVIDD843 pKa = 4.05PVPVVITQNIVIQLDD858 pKa = 3.72DD859 pKa = 4.13NGTVVIDD866 pKa = 4.16PLQIDD871 pKa = 4.14NGSSSAVGIGSLSLDD886 pKa = 2.95ISAFNCEE893 pKa = 4.91DD894 pKa = 3.06INNPVTVTLTVTSTLGSSSTGTAIVTVQDD923 pKa = 4.35TIAPIAPILSDD934 pKa = 3.4ILGEE938 pKa = 4.32CSATATTPTTTDD950 pKa = 2.7NCAGSITGTTTDD962 pKa = 3.28SLEE965 pKa = 3.87YY966 pKa = 9.72TIQGTHH972 pKa = 6.87TITWSFDD979 pKa = 3.3DD980 pKa = 4.83GNGNTTTATQNVIVNDD996 pKa = 3.79TTAPIAITKK1005 pKa = 10.33NITIPLDD1012 pKa = 3.44ASGNAIITPQMINNGSSDD1030 pKa = 3.3NCEE1033 pKa = 3.68IDD1035 pKa = 5.75SITLDD1040 pKa = 3.29NTVFDD1045 pKa = 4.81CSNVDD1050 pKa = 4.98SSAQCNAGTALDD1062 pKa = 4.49LGADD1066 pKa = 3.46NDD1068 pKa = 4.04YY1069 pKa = 11.29VRR1071 pKa = 11.84IGDD1074 pKa = 3.91PFHH1077 pKa = 6.99EE1078 pKa = 5.16FSNQITVSWWVNFQGNSAWMTQSTFGMSSNNTNVWNMHH1116 pKa = 5.82ASGANGITWYY1126 pKa = 10.32INDD1129 pKa = 3.46NGAYY1133 pKa = 9.84RR1134 pKa = 11.84AVSLNTTAGWHH1145 pKa = 6.95LITGVADD1152 pKa = 3.7TTSTRR1157 pKa = 11.84LYY1159 pKa = 10.84LDD1161 pKa = 4.1GNLASTDD1168 pKa = 3.27TGINNIRR1175 pKa = 11.84VNANSEE1181 pKa = 3.74IAIGIDD1187 pKa = 3.17GRR1189 pKa = 11.84IPSRR1193 pKa = 11.84NRR1195 pKa = 11.84NFQTDD1200 pKa = 3.03EE1201 pKa = 3.72VLIYY1205 pKa = 10.87NRR1207 pKa = 11.84ALSEE1211 pKa = 4.27TEE1213 pKa = 3.88IQDD1216 pKa = 5.18LLMNCPDD1223 pKa = 3.4ISDD1226 pKa = 3.87NSLVGYY1232 pKa = 9.13WAMNEE1237 pKa = 4.05GSGNTIEE1244 pKa = 4.74DD1245 pKa = 3.53QSITGANGVLINMTDD1260 pKa = 3.79ADD1262 pKa = 4.18WVTSGINNIGAGSSNEE1278 pKa = 3.83VTLTVTDD1285 pKa = 3.74VNGNSSTAIANVTVNDD1301 pKa = 3.33ITAPTLNDD1309 pKa = 2.9VPANVTVEE1317 pKa = 4.06CDD1319 pKa = 4.2AIPNAATISASDD1331 pKa = 3.36NCGNPTITFVEE1342 pKa = 4.06EE1343 pKa = 4.47RR1344 pKa = 11.84IDD1346 pKa = 4.12GKK1348 pKa = 11.74SNDD1351 pKa = 3.61NYY1353 pKa = 10.36TLNRR1357 pKa = 11.84TWTATDD1363 pKa = 3.2QSEE1366 pKa = 4.21NSVSHH1371 pKa = 4.98TQIITVQDD1379 pKa = 3.28TTAPFFDD1386 pKa = 4.3ATPVDD1391 pKa = 3.37ITVEE1395 pKa = 4.23CEE1397 pKa = 4.11TVPAAATISASDD1409 pKa = 3.36NCGNPTITFVEE1420 pKa = 4.15EE1421 pKa = 4.48RR1422 pKa = 11.84IDD1424 pKa = 5.03GISNDD1429 pKa = 3.53NYY1431 pKa = 10.17TLTRR1435 pKa = 11.84TWTATDD1441 pKa = 3.92LSGNTVSEE1449 pKa = 4.28SQIITVQDD1457 pKa = 3.22VTAPTVVGQNISVTLTSTGTVSIVPEE1483 pKa = 3.92NVLDD1487 pKa = 4.39NGSDD1491 pKa = 3.26NCGTVTYY1498 pKa = 10.24SVSQDD1503 pKa = 2.96TFGATDD1509 pKa = 3.57AVNSPVTVQLTGTDD1523 pKa = 3.24ASGNATTVPVLVTVIDD1539 pKa = 4.01PVPVVITQNIVVQLDD1554 pKa = 3.65DD1555 pKa = 4.12NGTVVIDD1562 pKa = 4.31PSQIDD1567 pKa = 3.71NGSSSAVGIGSLSLDD1582 pKa = 2.95ISAFNCEE1589 pKa = 4.91DD1590 pKa = 3.06INNPVTVTLTVTSTLGSSSTGTAIVTVQDD1619 pKa = 4.35TIAPIAPILSDD1630 pKa = 3.4ILGEE1634 pKa = 4.32CSATATTPTTTDD1646 pKa = 2.7NCAGSITGTTTDD1658 pKa = 3.66YY1659 pKa = 11.67LEE1661 pKa = 4.18YY1662 pKa = 8.84TAQGIHH1668 pKa = 6.29VITWSFDD1675 pKa = 3.44DD1676 pKa = 4.34GNGNIATATQNVIVIDD1692 pKa = 3.78VTAPTAIAQDD1702 pKa = 3.25ISINLDD1708 pKa = 3.4DD1709 pKa = 5.21NGNATITPQQIDD1721 pKa = 3.68NDD1723 pKa = 4.42SNDD1726 pKa = 3.21ACGIASTSLDD1736 pKa = 3.95KK1737 pKa = 11.66NSFDD1741 pKa = 4.0CSNVGSSASSSSFSFDD1757 pKa = 3.76GIANANGDD1765 pKa = 4.38EE1766 pKa = 4.1ILVNDD1771 pKa = 4.38TSFSTLTAQTIEE1783 pKa = 3.66AWIRR1787 pKa = 11.84PASGSNGNEE1796 pKa = 3.24SWLAIRR1802 pKa = 11.84DD1803 pKa = 3.56RR1804 pKa = 11.84VGNSRR1809 pKa = 11.84YY1810 pKa = 10.11SIHH1813 pKa = 6.77TNPSANTIGVYY1824 pKa = 10.58NSTHH1828 pKa = 6.72GYY1830 pKa = 7.49RR1831 pKa = 11.84TISISTGINANQWYY1845 pKa = 8.62HH1846 pKa = 5.27VAMVMNTTRR1855 pKa = 11.84TLVYY1859 pKa = 11.04VNGTLAGQIAAGISTNHH1876 pKa = 6.56GANVPLSIGSPNDD1889 pKa = 3.02ASYY1892 pKa = 11.4YY1893 pKa = 8.15GEE1895 pKa = 4.09KK1896 pKa = 9.11WQGDD1900 pKa = 3.83MDD1902 pKa = 5.85DD1903 pKa = 3.8LRR1905 pKa = 11.84VWNTEE1910 pKa = 3.45RR1911 pKa = 11.84TATEE1915 pKa = 3.55IADD1918 pKa = 3.58NRR1920 pKa = 11.84NNEE1923 pKa = 3.97LTGNEE1928 pKa = 4.04VGLVGYY1934 pKa = 9.75FNFNEE1939 pKa = 4.43GSGNSVDD1946 pKa = 4.29NLAGTINGTANNSQTEE1962 pKa = 3.99NWIDD1966 pKa = 3.59EE1967 pKa = 4.48SPISGSTGNMVTLTVTDD1984 pKa = 3.69VNGNSSTATANVTVVDD2000 pKa = 4.61NISPEE2005 pKa = 3.82ISDD2008 pKa = 3.81PADD2011 pKa = 3.04INVFATSAAGAVVNYY2026 pKa = 6.75NTPIGTDD2033 pKa = 2.93NCTVTTALTAGLADD2047 pKa = 4.27GEE2049 pKa = 4.6TFPIGTTVVSYY2060 pKa = 8.75TATDD2064 pKa = 3.93GSGNTATATFNVIVTGLPPEE2084 pKa = 4.1IVVPEE2089 pKa = 5.1DD2090 pKa = 2.9ITVNNDD2096 pKa = 2.76SGEE2099 pKa = 4.17CGAIVNYY2106 pKa = 10.33AATEE2110 pKa = 4.26TVGIPASTITYY2121 pKa = 9.62DD2122 pKa = 3.37IEE2124 pKa = 4.8PGSFFAEE2131 pKa = 4.37GTVTVTATATNPVGTSTKK2149 pKa = 9.99TFTVTVNDD2157 pKa = 3.56NQLPSVLTQNLIINLDD2173 pKa = 3.66ANGNSTITPQQIDD2186 pKa = 3.48NGSSDD2191 pKa = 3.35NCGIASISLDD2201 pKa = 3.17NDD2203 pKa = 3.54TFNCSNVSTNTVTLTVTDD2221 pKa = 3.58VNGNSNSLTANVTVNDD2237 pKa = 3.37VTAPNVITQNIIIEE2251 pKa = 4.09LDD2253 pKa = 3.62EE2254 pKa = 4.78NGEE2257 pKa = 4.01ASITPADD2264 pKa = 3.7IDD2266 pKa = 4.01NNSTDD2271 pKa = 2.92ACGIEE2276 pKa = 4.97SYY2278 pKa = 11.34SLDD2281 pKa = 3.99KK2282 pKa = 11.44DD2283 pKa = 4.41SFDD2286 pKa = 5.21CSNTGDD2292 pKa = 3.63NTVVLTVTDD2301 pKa = 3.68VNGNDD2306 pKa = 3.45ASLSATVTVQDD2317 pKa = 4.41NIAPTVITQSYY2328 pKa = 10.47SIDD2331 pKa = 3.7LTNGVADD2338 pKa = 5.34IIPSNIDD2345 pKa = 2.9GGTFDD2350 pKa = 4.65NCDD2353 pKa = 3.19FTLSIDD2359 pKa = 3.87RR2360 pKa = 11.84DD2361 pKa = 3.81HH2362 pKa = 6.59FTCDD2366 pKa = 3.82DD2367 pKa = 3.63IGDD2370 pKa = 4.09HH2371 pKa = 6.21VVTLTATDD2379 pKa = 3.49ASGNSSSEE2387 pKa = 3.98TAIVSILGNVPTIGINDD2404 pKa = 3.57FNIVQTQKK2412 pKa = 11.22KK2413 pKa = 7.03NTIFLGFGPQSINLSTVVDD2432 pKa = 4.16GGSGFTYY2439 pKa = 10.38EE2440 pKa = 4.12WTTSTGEE2447 pKa = 4.27VVSNQANPTINPTVSTTYY2465 pKa = 10.86YY2466 pKa = 9.36VTVTNANGCTASTSIYY2482 pKa = 10.2VCVIDD2487 pKa = 4.8ARR2489 pKa = 11.84AYY2491 pKa = 9.95DD2492 pKa = 3.28RR2493 pKa = 11.84KK2494 pKa = 10.5GRR2496 pKa = 11.84YY2497 pKa = 7.95KK2498 pKa = 10.86GKK2500 pKa = 10.51VLVCHH2505 pKa = 5.99HH2506 pKa = 6.69TNGKK2510 pKa = 9.92KK2511 pKa = 8.63GTKK2514 pKa = 9.44HH2515 pKa = 5.3VMISISKK2522 pKa = 8.21NAVMQHH2528 pKa = 5.13LTKK2531 pKa = 10.67HH2532 pKa = 6.45GIGTDD2537 pKa = 3.36HH2538 pKa = 7.49ADD2540 pKa = 4.06SLGACNAQCVTNQTILARR2558 pKa = 11.84TKK2560 pKa = 9.87TINLSSVTTYY2570 pKa = 10.43PNPSNGIFEE2579 pKa = 4.38VRR2581 pKa = 11.84LSKK2584 pKa = 11.4AEE2586 pKa = 4.21DD2587 pKa = 3.33NTRR2590 pKa = 11.84LLLFDD2595 pKa = 4.2LTGKK2599 pKa = 10.77LILHH2603 pKa = 5.89KK2604 pKa = 9.7TISTKK2609 pKa = 9.9KK2610 pKa = 10.79ASMGSNNLPSGVYY2623 pKa = 8.69ILKK2626 pKa = 10.52VIYY2629 pKa = 10.2NDD2631 pKa = 3.99SISTKK2636 pKa = 10.53KK2637 pKa = 10.84LIIEE2641 pKa = 4.28KK2642 pKa = 10.67GKK2644 pKa = 10.69
MM1 pKa = 7.48KK2 pKa = 10.52NNYY5 pKa = 9.87SKK7 pKa = 11.13FILKK11 pKa = 8.86TLLLFAILFGLEE23 pKa = 3.88TYY25 pKa = 10.01AQQNIIAQTQNITVQLDD42 pKa = 3.56ANGEE46 pKa = 4.25AIITPQQINNGSTATTGIQSLNLDD70 pKa = 3.62TSDD73 pKa = 5.29FNCTDD78 pKa = 2.94INGDD82 pKa = 3.69RR83 pKa = 11.84NFSYY87 pKa = 11.0LLDD90 pKa = 4.5GEE92 pKa = 4.49TDD94 pKa = 3.23ATGDD98 pKa = 4.04EE99 pKa = 4.34ILVDD103 pKa = 3.93DD104 pKa = 4.79TNLTTLTTQTIEE116 pKa = 3.44AWIRR120 pKa = 11.84PATGASDD127 pKa = 4.0NDD129 pKa = 3.31VLVAVRR135 pKa = 11.84NNIHH139 pKa = 6.24NSRR142 pKa = 11.84FSIHH146 pKa = 6.62INADD150 pKa = 3.29TNTIGIFNSTNWQTITTIINANQWYY175 pKa = 9.43HH176 pKa = 6.08IALVMNSTKK185 pKa = 10.36TLVYY189 pKa = 10.51LDD191 pKa = 3.92GAFEE195 pKa = 4.68GEE197 pKa = 3.9ISTGINTSLGANAPLSIGSPNDD219 pKa = 3.44TGWYY223 pKa = 7.61SEE225 pKa = 3.87KK226 pKa = 10.41WEE228 pKa = 4.36GNIDD232 pKa = 5.46DD233 pKa = 4.21LRR235 pKa = 11.84IWNDD239 pKa = 2.44EE240 pKa = 3.95RR241 pKa = 11.84TALEE245 pKa = 3.64IANNRR250 pKa = 11.84NIEE253 pKa = 4.11LVGNEE258 pKa = 4.22AGLVGYY264 pKa = 10.48FDD266 pKa = 5.05FNEE269 pKa = 4.58GTGNTITNLAGTVNGTANNSQNANWSTDD297 pKa = 3.14SPIEE301 pKa = 4.42SNGNTVNLTVTDD313 pKa = 3.86VNGNSATTPANVIVVDD329 pKa = 5.01NIAPIAPTLADD340 pKa = 3.43VTGEE344 pKa = 4.02CSATVTAPTTTDD356 pKa = 2.77NCAGTITGTTTDD368 pKa = 3.32SLTYY372 pKa = 7.42TTQGTHH378 pKa = 6.68IITWNFDD385 pKa = 3.58DD386 pKa = 5.63GNGNSTTASQNVIISDD402 pKa = 3.98STPPTAITNNISVNLDD418 pKa = 3.23ANGNASITPQQIDD431 pKa = 3.55NGSSDD436 pKa = 3.43ACGISSISCSRR447 pKa = 11.84VGGSGNGLTNFLNGTLNNGTADD469 pKa = 3.21IIEE472 pKa = 4.33YY473 pKa = 10.71SNSNAPNYY481 pKa = 7.41RR482 pKa = 11.84TSFGTTCNEE491 pKa = 3.02AWFARR496 pKa = 11.84GMLCDD501 pKa = 4.66LAPNQNDD508 pKa = 3.73PDD510 pKa = 3.71NSSRR514 pKa = 11.84QDD516 pKa = 3.41NNAGATWRR524 pKa = 11.84NTSPGNSYY532 pKa = 10.56GRR534 pKa = 11.84LVIDD538 pKa = 4.97LGSSQTINAMSIFQMFSDD556 pKa = 4.19GKK558 pKa = 6.81TTHH561 pKa = 6.72IEE563 pKa = 3.62AFAYY567 pKa = 10.21NGNLEE572 pKa = 4.36NPNSSSPGWNQLFPSTSVGAGVLNGTTVSSPLKK605 pKa = 8.72TTFQSTNTRR614 pKa = 11.84FIKK617 pKa = 10.58LYY619 pKa = 10.44AKK621 pKa = 10.52NDD623 pKa = 3.28GSLGFSGYY631 pKa = 9.92IEE633 pKa = 4.04LRR635 pKa = 11.84QVKK638 pKa = 8.98VFNTNEE644 pKa = 3.77SNACDD649 pKa = 4.49FNCDD653 pKa = 3.79DD654 pKa = 4.63LGEE657 pKa = 4.41NNVTLTVEE665 pKa = 4.49DD666 pKa = 3.87KK667 pKa = 11.19NGNISTKK674 pKa = 10.5DD675 pKa = 3.51AIVTIIDD682 pKa = 4.82AIAPALEE689 pKa = 4.32ATPADD694 pKa = 3.89LTVEE698 pKa = 4.25CHH700 pKa = 6.73SIPDD704 pKa = 3.56VATISATDD712 pKa = 3.38NCDD715 pKa = 3.33SNISVNFTEE724 pKa = 4.09QRR726 pKa = 11.84ADD728 pKa = 3.23GFSNDD733 pKa = 3.57NYY735 pKa = 10.28TLTRR739 pKa = 11.84TWSATDD745 pKa = 3.21QSGNTVSEE753 pKa = 4.39SQVITVQDD761 pKa = 3.69VTPPTIVGQNISVTLTASGTVSIIPQNVLNNGSDD795 pKa = 3.37TCSTVSYY802 pKa = 9.46TISQNTFGATDD813 pKa = 5.5AINSPVTIQLIGTDD827 pKa = 3.51EE828 pKa = 4.43SGNATTIPVQVTVIDD843 pKa = 4.05PVPVVITQNIVIQLDD858 pKa = 3.72DD859 pKa = 4.13NGTVVIDD866 pKa = 4.16PLQIDD871 pKa = 4.14NGSSSAVGIGSLSLDD886 pKa = 2.95ISAFNCEE893 pKa = 4.91DD894 pKa = 3.06INNPVTVTLTVTSTLGSSSTGTAIVTVQDD923 pKa = 4.35TIAPIAPILSDD934 pKa = 3.4ILGEE938 pKa = 4.32CSATATTPTTTDD950 pKa = 2.7NCAGSITGTTTDD962 pKa = 3.28SLEE965 pKa = 3.87YY966 pKa = 9.72TIQGTHH972 pKa = 6.87TITWSFDD979 pKa = 3.3DD980 pKa = 4.83GNGNTTTATQNVIVNDD996 pKa = 3.79TTAPIAITKK1005 pKa = 10.33NITIPLDD1012 pKa = 3.44ASGNAIITPQMINNGSSDD1030 pKa = 3.3NCEE1033 pKa = 3.68IDD1035 pKa = 5.75SITLDD1040 pKa = 3.29NTVFDD1045 pKa = 4.81CSNVDD1050 pKa = 4.98SSAQCNAGTALDD1062 pKa = 4.49LGADD1066 pKa = 3.46NDD1068 pKa = 4.04YY1069 pKa = 11.29VRR1071 pKa = 11.84IGDD1074 pKa = 3.91PFHH1077 pKa = 6.99EE1078 pKa = 5.16FSNQITVSWWVNFQGNSAWMTQSTFGMSSNNTNVWNMHH1116 pKa = 5.82ASGANGITWYY1126 pKa = 10.32INDD1129 pKa = 3.46NGAYY1133 pKa = 9.84RR1134 pKa = 11.84AVSLNTTAGWHH1145 pKa = 6.95LITGVADD1152 pKa = 3.7TTSTRR1157 pKa = 11.84LYY1159 pKa = 10.84LDD1161 pKa = 4.1GNLASTDD1168 pKa = 3.27TGINNIRR1175 pKa = 11.84VNANSEE1181 pKa = 3.74IAIGIDD1187 pKa = 3.17GRR1189 pKa = 11.84IPSRR1193 pKa = 11.84NRR1195 pKa = 11.84NFQTDD1200 pKa = 3.03EE1201 pKa = 3.72VLIYY1205 pKa = 10.87NRR1207 pKa = 11.84ALSEE1211 pKa = 4.27TEE1213 pKa = 3.88IQDD1216 pKa = 5.18LLMNCPDD1223 pKa = 3.4ISDD1226 pKa = 3.87NSLVGYY1232 pKa = 9.13WAMNEE1237 pKa = 4.05GSGNTIEE1244 pKa = 4.74DD1245 pKa = 3.53QSITGANGVLINMTDD1260 pKa = 3.79ADD1262 pKa = 4.18WVTSGINNIGAGSSNEE1278 pKa = 3.83VTLTVTDD1285 pKa = 3.74VNGNSSTAIANVTVNDD1301 pKa = 3.33ITAPTLNDD1309 pKa = 2.9VPANVTVEE1317 pKa = 4.06CDD1319 pKa = 4.2AIPNAATISASDD1331 pKa = 3.36NCGNPTITFVEE1342 pKa = 4.06EE1343 pKa = 4.47RR1344 pKa = 11.84IDD1346 pKa = 4.12GKK1348 pKa = 11.74SNDD1351 pKa = 3.61NYY1353 pKa = 10.36TLNRR1357 pKa = 11.84TWTATDD1363 pKa = 3.2QSEE1366 pKa = 4.21NSVSHH1371 pKa = 4.98TQIITVQDD1379 pKa = 3.28TTAPFFDD1386 pKa = 4.3ATPVDD1391 pKa = 3.37ITVEE1395 pKa = 4.23CEE1397 pKa = 4.11TVPAAATISASDD1409 pKa = 3.36NCGNPTITFVEE1420 pKa = 4.15EE1421 pKa = 4.48RR1422 pKa = 11.84IDD1424 pKa = 5.03GISNDD1429 pKa = 3.53NYY1431 pKa = 10.17TLTRR1435 pKa = 11.84TWTATDD1441 pKa = 3.92LSGNTVSEE1449 pKa = 4.28SQIITVQDD1457 pKa = 3.22VTAPTVVGQNISVTLTSTGTVSIVPEE1483 pKa = 3.92NVLDD1487 pKa = 4.39NGSDD1491 pKa = 3.26NCGTVTYY1498 pKa = 10.24SVSQDD1503 pKa = 2.96TFGATDD1509 pKa = 3.57AVNSPVTVQLTGTDD1523 pKa = 3.24ASGNATTVPVLVTVIDD1539 pKa = 4.01PVPVVITQNIVVQLDD1554 pKa = 3.65DD1555 pKa = 4.12NGTVVIDD1562 pKa = 4.31PSQIDD1567 pKa = 3.71NGSSSAVGIGSLSLDD1582 pKa = 2.95ISAFNCEE1589 pKa = 4.91DD1590 pKa = 3.06INNPVTVTLTVTSTLGSSSTGTAIVTVQDD1619 pKa = 4.35TIAPIAPILSDD1630 pKa = 3.4ILGEE1634 pKa = 4.32CSATATTPTTTDD1646 pKa = 2.7NCAGSITGTTTDD1658 pKa = 3.66YY1659 pKa = 11.67LEE1661 pKa = 4.18YY1662 pKa = 8.84TAQGIHH1668 pKa = 6.29VITWSFDD1675 pKa = 3.44DD1676 pKa = 4.34GNGNIATATQNVIVIDD1692 pKa = 3.78VTAPTAIAQDD1702 pKa = 3.25ISINLDD1708 pKa = 3.4DD1709 pKa = 5.21NGNATITPQQIDD1721 pKa = 3.68NDD1723 pKa = 4.42SNDD1726 pKa = 3.21ACGIASTSLDD1736 pKa = 3.95KK1737 pKa = 11.66NSFDD1741 pKa = 4.0CSNVGSSASSSSFSFDD1757 pKa = 3.76GIANANGDD1765 pKa = 4.38EE1766 pKa = 4.1ILVNDD1771 pKa = 4.38TSFSTLTAQTIEE1783 pKa = 3.66AWIRR1787 pKa = 11.84PASGSNGNEE1796 pKa = 3.24SWLAIRR1802 pKa = 11.84DD1803 pKa = 3.56RR1804 pKa = 11.84VGNSRR1809 pKa = 11.84YY1810 pKa = 10.11SIHH1813 pKa = 6.77TNPSANTIGVYY1824 pKa = 10.58NSTHH1828 pKa = 6.72GYY1830 pKa = 7.49RR1831 pKa = 11.84TISISTGINANQWYY1845 pKa = 8.62HH1846 pKa = 5.27VAMVMNTTRR1855 pKa = 11.84TLVYY1859 pKa = 11.04VNGTLAGQIAAGISTNHH1876 pKa = 6.56GANVPLSIGSPNDD1889 pKa = 3.02ASYY1892 pKa = 11.4YY1893 pKa = 8.15GEE1895 pKa = 4.09KK1896 pKa = 9.11WQGDD1900 pKa = 3.83MDD1902 pKa = 5.85DD1903 pKa = 3.8LRR1905 pKa = 11.84VWNTEE1910 pKa = 3.45RR1911 pKa = 11.84TATEE1915 pKa = 3.55IADD1918 pKa = 3.58NRR1920 pKa = 11.84NNEE1923 pKa = 3.97LTGNEE1928 pKa = 4.04VGLVGYY1934 pKa = 9.75FNFNEE1939 pKa = 4.43GSGNSVDD1946 pKa = 4.29NLAGTINGTANNSQTEE1962 pKa = 3.99NWIDD1966 pKa = 3.59EE1967 pKa = 4.48SPISGSTGNMVTLTVTDD1984 pKa = 3.69VNGNSSTATANVTVVDD2000 pKa = 4.61NISPEE2005 pKa = 3.82ISDD2008 pKa = 3.81PADD2011 pKa = 3.04INVFATSAAGAVVNYY2026 pKa = 6.75NTPIGTDD2033 pKa = 2.93NCTVTTALTAGLADD2047 pKa = 4.27GEE2049 pKa = 4.6TFPIGTTVVSYY2060 pKa = 8.75TATDD2064 pKa = 3.93GSGNTATATFNVIVTGLPPEE2084 pKa = 4.1IVVPEE2089 pKa = 5.1DD2090 pKa = 2.9ITVNNDD2096 pKa = 2.76SGEE2099 pKa = 4.17CGAIVNYY2106 pKa = 10.33AATEE2110 pKa = 4.26TVGIPASTITYY2121 pKa = 9.62DD2122 pKa = 3.37IEE2124 pKa = 4.8PGSFFAEE2131 pKa = 4.37GTVTVTATATNPVGTSTKK2149 pKa = 9.99TFTVTVNDD2157 pKa = 3.56NQLPSVLTQNLIINLDD2173 pKa = 3.66ANGNSTITPQQIDD2186 pKa = 3.48NGSSDD2191 pKa = 3.35NCGIASISLDD2201 pKa = 3.17NDD2203 pKa = 3.54TFNCSNVSTNTVTLTVTDD2221 pKa = 3.58VNGNSNSLTANVTVNDD2237 pKa = 3.37VTAPNVITQNIIIEE2251 pKa = 4.09LDD2253 pKa = 3.62EE2254 pKa = 4.78NGEE2257 pKa = 4.01ASITPADD2264 pKa = 3.7IDD2266 pKa = 4.01NNSTDD2271 pKa = 2.92ACGIEE2276 pKa = 4.97SYY2278 pKa = 11.34SLDD2281 pKa = 3.99KK2282 pKa = 11.44DD2283 pKa = 4.41SFDD2286 pKa = 5.21CSNTGDD2292 pKa = 3.63NTVVLTVTDD2301 pKa = 3.68VNGNDD2306 pKa = 3.45ASLSATVTVQDD2317 pKa = 4.41NIAPTVITQSYY2328 pKa = 10.47SIDD2331 pKa = 3.7LTNGVADD2338 pKa = 5.34IIPSNIDD2345 pKa = 2.9GGTFDD2350 pKa = 4.65NCDD2353 pKa = 3.19FTLSIDD2359 pKa = 3.87RR2360 pKa = 11.84DD2361 pKa = 3.81HH2362 pKa = 6.59FTCDD2366 pKa = 3.82DD2367 pKa = 3.63IGDD2370 pKa = 4.09HH2371 pKa = 6.21VVTLTATDD2379 pKa = 3.49ASGNSSSEE2387 pKa = 3.98TAIVSILGNVPTIGINDD2404 pKa = 3.57FNIVQTQKK2412 pKa = 11.22KK2413 pKa = 7.03NTIFLGFGPQSINLSTVVDD2432 pKa = 4.16GGSGFTYY2439 pKa = 10.38EE2440 pKa = 4.12WTTSTGEE2447 pKa = 4.27VVSNQANPTINPTVSTTYY2465 pKa = 10.86YY2466 pKa = 9.36VTVTNANGCTASTSIYY2482 pKa = 10.2VCVIDD2487 pKa = 4.8ARR2489 pKa = 11.84AYY2491 pKa = 9.95DD2492 pKa = 3.28RR2493 pKa = 11.84KK2494 pKa = 10.5GRR2496 pKa = 11.84YY2497 pKa = 7.95KK2498 pKa = 10.86GKK2500 pKa = 10.51VLVCHH2505 pKa = 5.99HH2506 pKa = 6.69TNGKK2510 pKa = 9.92KK2511 pKa = 8.63GTKK2514 pKa = 9.44HH2515 pKa = 5.3VMISISKK2522 pKa = 8.21NAVMQHH2528 pKa = 5.13LTKK2531 pKa = 10.67HH2532 pKa = 6.45GIGTDD2537 pKa = 3.36HH2538 pKa = 7.49ADD2540 pKa = 4.06SLGACNAQCVTNQTILARR2558 pKa = 11.84TKK2560 pKa = 9.87TINLSSVTTYY2570 pKa = 10.43PNPSNGIFEE2579 pKa = 4.38VRR2581 pKa = 11.84LSKK2584 pKa = 11.4AEE2586 pKa = 4.21DD2587 pKa = 3.33NTRR2590 pKa = 11.84LLLFDD2595 pKa = 4.2LTGKK2599 pKa = 10.77LILHH2603 pKa = 5.89KK2604 pKa = 9.7TISTKK2609 pKa = 9.9KK2610 pKa = 10.79ASMGSNNLPSGVYY2623 pKa = 8.69ILKK2626 pKa = 10.52VIYY2629 pKa = 10.2NDD2631 pKa = 3.99SISTKK2636 pKa = 10.53KK2637 pKa = 10.84LIIEE2641 pKa = 4.28KK2642 pKa = 10.67GKK2644 pKa = 10.69
Molecular weight: 277.62 kDa
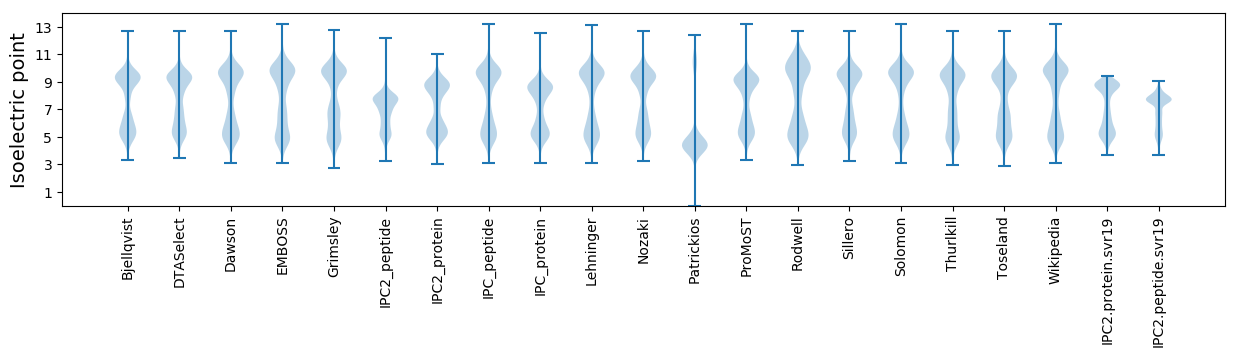
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A559S2L1|A0A559S2L1_9FLAO NTE family protein OS=Lutibacter sp. Hel_I_33_5 OX=1566289 GN=OD91_1881 PE=4 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.34KK6 pKa = 9.56RR7 pKa = 11.84KK8 pKa = 8.34RR9 pKa = 11.84AKK11 pKa = 10.25ISTHH15 pKa = 4.55KK16 pKa = 9.9RR17 pKa = 11.84KK18 pKa = 9.85KK19 pKa = 8.28RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.7KK27 pKa = 10.78KK28 pKa = 9.39KK29 pKa = 10.83
MM1 pKa = 7.84PSGKK5 pKa = 9.34KK6 pKa = 9.56RR7 pKa = 11.84KK8 pKa = 8.34RR9 pKa = 11.84AKK11 pKa = 10.25ISTHH15 pKa = 4.55KK16 pKa = 9.9RR17 pKa = 11.84KK18 pKa = 9.85KK19 pKa = 8.28RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.7KK27 pKa = 10.78KK28 pKa = 9.39KK29 pKa = 10.83
Molecular weight: 3.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
952811 |
29 |
9603 |
361.7 |
40.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.861 ± 0.064 | 0.705 ± 0.017 |
5.787 ± 0.181 | 6.153 ± 0.08 |
5.327 ± 0.082 | 6.394 ± 0.097 |
1.607 ± 0.03 | 8.418 ± 0.084 |
8.647 ± 0.152 | 8.935 ± 0.087 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.872 ± 0.034 | 7.012 ± 0.068 |
3.128 ± 0.046 | 3.042 ± 0.035 |
3.002 ± 0.059 | 6.817 ± 0.054 |
6.335 ± 0.186 | 6.103 ± 0.049 |
0.962 ± 0.019 | 3.895 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
