
Capybara microvirus Cap3_SP_550
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.43
Get precalculated fractions of proteins
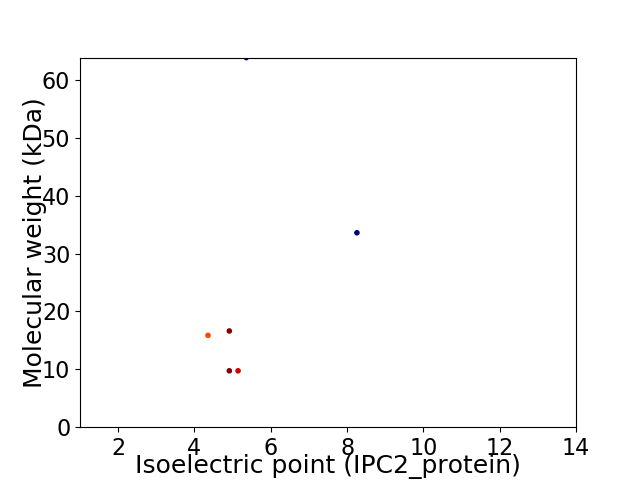
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W532|A0A4P8W532_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_550 OX=2585474 PE=4 SV=1
MM1 pKa = 6.96AQKK4 pKa = 10.4IIDD7 pKa = 3.61TDD9 pKa = 3.95GVVNKK14 pKa = 9.95LPEE17 pKa = 4.49YY18 pKa = 7.6GTPGYY23 pKa = 10.15IFEE26 pKa = 4.92LAWHH30 pKa = 7.18DD31 pKa = 3.97EE32 pKa = 4.01AKK34 pKa = 10.28KK35 pKa = 10.71DD36 pKa = 3.74YY37 pKa = 11.04LVVVDD42 pKa = 5.66AKK44 pKa = 11.22PEE46 pKa = 4.09VEE48 pKa = 4.5VMNARR53 pKa = 11.84AKK55 pKa = 10.78GVTPYY60 pKa = 11.38DD61 pKa = 4.31LISTFGGIDD70 pKa = 3.38EE71 pKa = 4.38VTQAFASKK79 pKa = 9.73EE80 pKa = 3.93ASAVYY85 pKa = 10.55ADD87 pKa = 3.68VSGVPEE93 pKa = 4.13FASDD97 pKa = 3.62DD98 pKa = 4.03VYY100 pKa = 11.82DD101 pKa = 3.96SVIARR106 pKa = 11.84LKK108 pKa = 10.88ADD110 pKa = 3.15IAAYY114 pKa = 8.09EE115 pKa = 4.1ASHH118 pKa = 6.67AKK120 pKa = 9.28TDD122 pKa = 3.66EE123 pKa = 4.25SSKK126 pKa = 9.4EE127 pKa = 3.87TAEE130 pKa = 4.14EE131 pKa = 3.69VDD133 pKa = 3.45NEE135 pKa = 4.26VNKK138 pKa = 10.17EE139 pKa = 4.13VNKK142 pKa = 10.93GEE144 pKa = 4.35SKK146 pKa = 10.99
MM1 pKa = 6.96AQKK4 pKa = 10.4IIDD7 pKa = 3.61TDD9 pKa = 3.95GVVNKK14 pKa = 9.95LPEE17 pKa = 4.49YY18 pKa = 7.6GTPGYY23 pKa = 10.15IFEE26 pKa = 4.92LAWHH30 pKa = 7.18DD31 pKa = 3.97EE32 pKa = 4.01AKK34 pKa = 10.28KK35 pKa = 10.71DD36 pKa = 3.74YY37 pKa = 11.04LVVVDD42 pKa = 5.66AKK44 pKa = 11.22PEE46 pKa = 4.09VEE48 pKa = 4.5VMNARR53 pKa = 11.84AKK55 pKa = 10.78GVTPYY60 pKa = 11.38DD61 pKa = 4.31LISTFGGIDD70 pKa = 3.38EE71 pKa = 4.38VTQAFASKK79 pKa = 9.73EE80 pKa = 3.93ASAVYY85 pKa = 10.55ADD87 pKa = 3.68VSGVPEE93 pKa = 4.13FASDD97 pKa = 3.62DD98 pKa = 4.03VYY100 pKa = 11.82DD101 pKa = 3.96SVIARR106 pKa = 11.84LKK108 pKa = 10.88ADD110 pKa = 3.15IAAYY114 pKa = 8.09EE115 pKa = 4.1ASHH118 pKa = 6.67AKK120 pKa = 9.28TDD122 pKa = 3.66EE123 pKa = 4.25SSKK126 pKa = 9.4EE127 pKa = 3.87TAEE130 pKa = 4.14EE131 pKa = 3.69VDD133 pKa = 3.45NEE135 pKa = 4.26VNKK138 pKa = 10.17EE139 pKa = 4.13VNKK142 pKa = 10.93GEE144 pKa = 4.35SKK146 pKa = 10.99
Molecular weight: 15.87 kDa
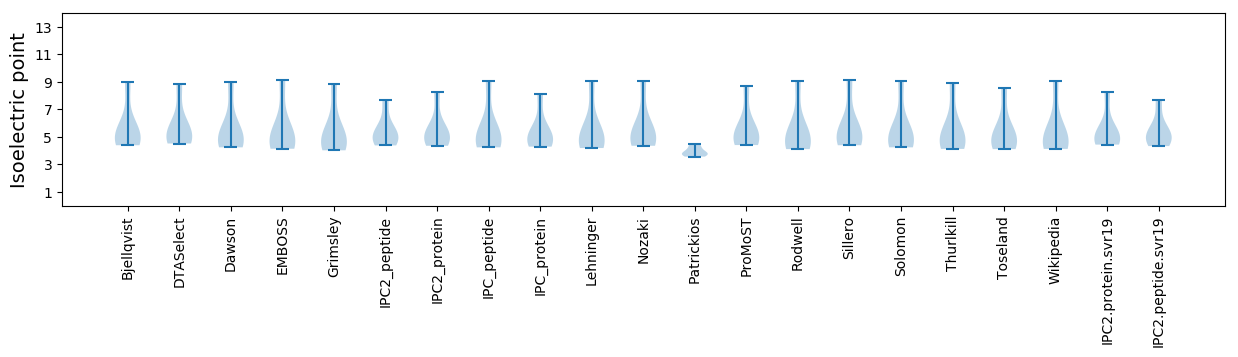
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W596|A0A4P8W596_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_550 OX=2585474 PE=4 SV=1
MM1 pKa = 6.97MCTRR5 pKa = 11.84PLHH8 pKa = 5.65YY9 pKa = 10.34FEE11 pKa = 6.71SNLIDD16 pKa = 3.7NEE18 pKa = 4.48TGKK21 pKa = 10.88KK22 pKa = 9.47PGFITGYY29 pKa = 7.1QVKK32 pKa = 9.99LAGFRR37 pKa = 11.84LFAKK41 pKa = 10.34HH42 pKa = 6.78GIACTEE48 pKa = 4.07TYY50 pKa = 9.73LAKK53 pKa = 10.19PVEE56 pKa = 4.75VPCGKK61 pKa = 10.37CPEE64 pKa = 4.13CRR66 pKa = 11.84KK67 pKa = 9.48ALKK70 pKa = 8.2MQWVARR76 pKa = 11.84AVAEE80 pKa = 4.46LEE82 pKa = 4.46TSKK85 pKa = 10.65HH86 pKa = 6.01AYY88 pKa = 9.85FLTLTYY94 pKa = 11.12DD95 pKa = 3.72NDD97 pKa = 4.02HH98 pKa = 6.75LPSDD102 pKa = 4.21FMCHH106 pKa = 6.35KK107 pKa = 10.29EE108 pKa = 4.06HH109 pKa = 6.77IQVFLNRR116 pKa = 11.84FRR118 pKa = 11.84KK119 pKa = 9.18NNKK122 pKa = 6.74TRR124 pKa = 11.84YY125 pKa = 9.53LIVGEE130 pKa = 4.29HH131 pKa = 6.74GEE133 pKa = 4.25LSNRR137 pKa = 11.84PHH139 pKa = 5.71YY140 pKa = 10.25HH141 pKa = 6.81AILYY145 pKa = 7.93TEE147 pKa = 4.4NPIDD151 pKa = 5.47DD152 pKa = 6.18LIPVNKK158 pKa = 10.29SKK160 pKa = 9.09TLPLYY165 pKa = 10.69EE166 pKa = 4.24SDD168 pKa = 4.53KK169 pKa = 10.49VTKK172 pKa = 9.52QWQNGIVKK180 pKa = 10.1IGVAYY185 pKa = 8.42PQAIAYY191 pKa = 8.97SVGYY195 pKa = 9.85LVSKK199 pKa = 9.74EE200 pKa = 3.84KK201 pKa = 9.73KK202 pKa = 8.26TAFKK206 pKa = 9.74MQSQGLGYY214 pKa = 10.51QFFSGLNRR222 pKa = 11.84RR223 pKa = 11.84YY224 pKa = 10.1VLSDD228 pKa = 2.83MHH230 pKa = 6.71GHH232 pKa = 5.54EE233 pKa = 5.34MIVSLPRR240 pKa = 11.84YY241 pKa = 8.93LKK243 pKa = 10.54KK244 pKa = 10.69KK245 pKa = 9.61YY246 pKa = 10.04DD247 pKa = 3.69LNFDD251 pKa = 3.86YY252 pKa = 10.95DD253 pKa = 3.99DD254 pKa = 4.38SLNSIKK260 pKa = 9.5WHH262 pKa = 6.0NEE264 pKa = 3.28VVASGLKK271 pKa = 10.15EE272 pKa = 3.49EE273 pKa = 4.57TYY275 pKa = 11.06RR276 pKa = 11.84DD277 pKa = 3.41FKK279 pKa = 11.36EE280 pKa = 3.84YY281 pKa = 11.06LEE283 pKa = 4.09EE284 pKa = 4.93SKK286 pKa = 10.8LQKK289 pKa = 10.62
MM1 pKa = 6.97MCTRR5 pKa = 11.84PLHH8 pKa = 5.65YY9 pKa = 10.34FEE11 pKa = 6.71SNLIDD16 pKa = 3.7NEE18 pKa = 4.48TGKK21 pKa = 10.88KK22 pKa = 9.47PGFITGYY29 pKa = 7.1QVKK32 pKa = 9.99LAGFRR37 pKa = 11.84LFAKK41 pKa = 10.34HH42 pKa = 6.78GIACTEE48 pKa = 4.07TYY50 pKa = 9.73LAKK53 pKa = 10.19PVEE56 pKa = 4.75VPCGKK61 pKa = 10.37CPEE64 pKa = 4.13CRR66 pKa = 11.84KK67 pKa = 9.48ALKK70 pKa = 8.2MQWVARR76 pKa = 11.84AVAEE80 pKa = 4.46LEE82 pKa = 4.46TSKK85 pKa = 10.65HH86 pKa = 6.01AYY88 pKa = 9.85FLTLTYY94 pKa = 11.12DD95 pKa = 3.72NDD97 pKa = 4.02HH98 pKa = 6.75LPSDD102 pKa = 4.21FMCHH106 pKa = 6.35KK107 pKa = 10.29EE108 pKa = 4.06HH109 pKa = 6.77IQVFLNRR116 pKa = 11.84FRR118 pKa = 11.84KK119 pKa = 9.18NNKK122 pKa = 6.74TRR124 pKa = 11.84YY125 pKa = 9.53LIVGEE130 pKa = 4.29HH131 pKa = 6.74GEE133 pKa = 4.25LSNRR137 pKa = 11.84PHH139 pKa = 5.71YY140 pKa = 10.25HH141 pKa = 6.81AILYY145 pKa = 7.93TEE147 pKa = 4.4NPIDD151 pKa = 5.47DD152 pKa = 6.18LIPVNKK158 pKa = 10.29SKK160 pKa = 9.09TLPLYY165 pKa = 10.69EE166 pKa = 4.24SDD168 pKa = 4.53KK169 pKa = 10.49VTKK172 pKa = 9.52QWQNGIVKK180 pKa = 10.1IGVAYY185 pKa = 8.42PQAIAYY191 pKa = 8.97SVGYY195 pKa = 9.85LVSKK199 pKa = 9.74EE200 pKa = 3.84KK201 pKa = 9.73KK202 pKa = 8.26TAFKK206 pKa = 9.74MQSQGLGYY214 pKa = 10.51QFFSGLNRR222 pKa = 11.84RR223 pKa = 11.84YY224 pKa = 10.1VLSDD228 pKa = 2.83MHH230 pKa = 6.71GHH232 pKa = 5.54EE233 pKa = 5.34MIVSLPRR240 pKa = 11.84YY241 pKa = 8.93LKK243 pKa = 10.54KK244 pKa = 10.69KK245 pKa = 9.61YY246 pKa = 10.04DD247 pKa = 3.69LNFDD251 pKa = 3.86YY252 pKa = 10.95DD253 pKa = 3.99DD254 pKa = 4.38SLNSIKK260 pKa = 9.5WHH262 pKa = 6.0NEE264 pKa = 3.28VVASGLKK271 pKa = 10.15EE272 pKa = 3.49EE273 pKa = 4.57TYY275 pKa = 11.06RR276 pKa = 11.84DD277 pKa = 3.41FKK279 pKa = 11.36EE280 pKa = 3.84YY281 pKa = 11.06LEE283 pKa = 4.09EE284 pKa = 4.93SKK286 pKa = 10.8LQKK289 pKa = 10.62
Molecular weight: 33.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1342 |
86 |
577 |
223.7 |
24.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.24 ± 1.407 | 1.043 ± 0.314 |
5.514 ± 0.719 | 6.855 ± 1.323 |
4.471 ± 0.558 | 6.408 ± 0.906 |
2.012 ± 0.525 | 5.44 ± 0.611 |
7.079 ± 1.471 | 7.899 ± 0.789 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.937 ± 0.22 | 5.365 ± 0.547 |
3.875 ± 0.765 | 2.906 ± 0.424 |
3.055 ± 0.662 | 7.973 ± 1.338 |
5.961 ± 0.644 | 6.334 ± 0.913 |
1.267 ± 0.351 | 5.365 ± 0.532 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
