
Natrialba magadii (strain ATCC 43099 / DSM 3394 / CCM 3739 / CIP 104546 / IAM 13178 / JCM 8861 / NBRC 102185 / NCIMB 2190 / MS3) (Natronobacterium magadii)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Natrialba; Natrialba magadii
Average proteome isoelectric point is 4.75
Get precalculated fractions of proteins
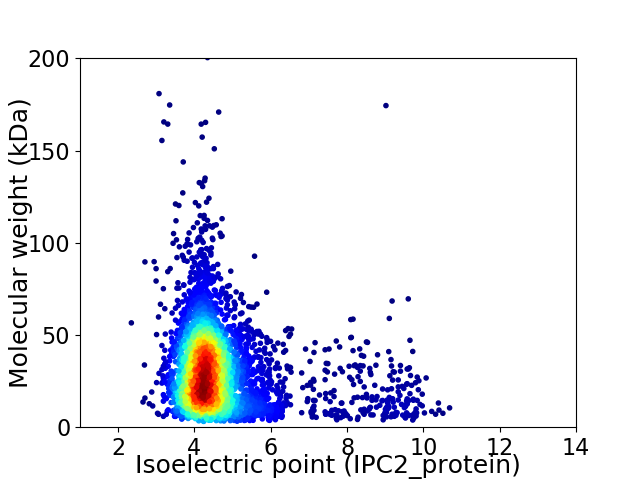
Virtual 2D-PAGE plot for 4203 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
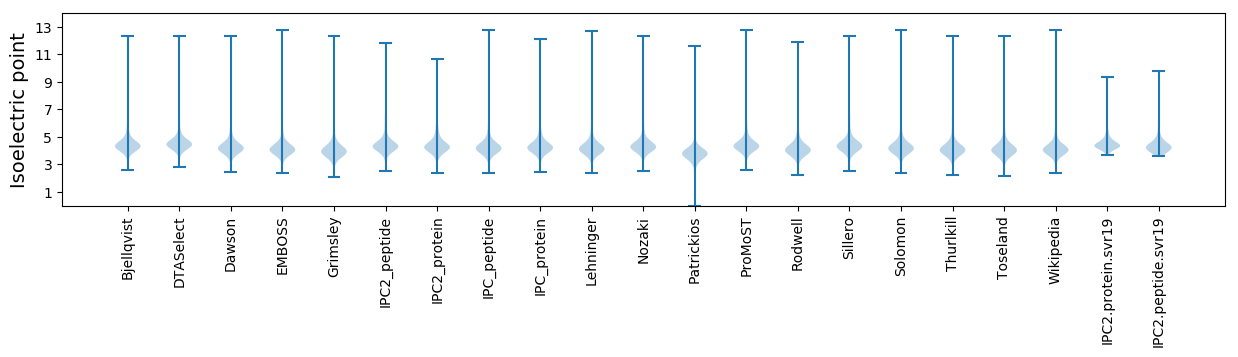
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D3SWB9|D3SWB9_NATMM Uncharacterized protein OS=Natrialba magadii (strain ATCC 43099 / DSM 3394 / CCM 3739 / CIP 104546 / IAM 13178 / JCM 8861 / NBRC 102185 / NCIMB 2190 / MS3) OX=547559 GN=Nmag_0113 PE=4 SV=1
MM1 pKa = 6.2VHH3 pKa = 6.53RR4 pKa = 11.84RR5 pKa = 11.84NVLKK9 pKa = 10.63GAGAISVAGMAGCLGGDD26 pKa = 3.52DD27 pKa = 4.58GEE29 pKa = 4.45YY30 pKa = 9.81TRR32 pKa = 11.84PVEE35 pKa = 4.93IDD37 pKa = 3.27FDD39 pKa = 3.82DD40 pKa = 4.27WPPEE44 pKa = 3.88EE45 pKa = 4.98YY46 pKa = 10.69GDD48 pKa = 5.77SLHH51 pKa = 6.28AWNWYY56 pKa = 9.36GEE58 pKa = 4.15WNEE61 pKa = 3.73WGAEE65 pKa = 4.04NFAEE69 pKa = 4.64EE70 pKa = 4.63YY71 pKa = 10.61DD72 pKa = 3.65LSSYY76 pKa = 7.42EE77 pKa = 4.18TEE79 pKa = 4.58VYY81 pKa = 7.94ATPSDD86 pKa = 3.43WFTNIQANPDD96 pKa = 3.08NHH98 pKa = 7.04GIDD101 pKa = 3.44HH102 pKa = 6.61VGLSTEE108 pKa = 4.04WIHH111 pKa = 7.22RR112 pKa = 11.84VNQEE116 pKa = 4.11DD117 pKa = 3.92ALEE120 pKa = 4.84PIPVDD125 pKa = 3.3EE126 pKa = 5.14LPNVDD131 pKa = 4.61VADD134 pKa = 3.9QYY136 pKa = 11.4MDD138 pKa = 3.0LHH140 pKa = 6.85RR141 pKa = 11.84EE142 pKa = 4.04HH143 pKa = 6.97FGSDD147 pKa = 3.18DD148 pKa = 3.76GVGGVYY154 pKa = 10.23AVPHH158 pKa = 6.89AIVLGPVVVYY168 pKa = 7.81NTNEE172 pKa = 3.77IQDD175 pKa = 4.05PPEE178 pKa = 5.91SIDD181 pKa = 3.47ILWDD185 pKa = 3.13EE186 pKa = 4.79EE187 pKa = 4.4YY188 pKa = 11.2ADD190 pKa = 5.12EE191 pKa = 5.07ISMMAHH197 pKa = 6.99LSGFLCEE204 pKa = 5.45AGALYY209 pKa = 9.84TGQDD213 pKa = 3.8PNDD216 pKa = 4.06PDD218 pKa = 3.78DD219 pKa = 4.35FEE221 pKa = 5.39EE222 pKa = 4.09IRR224 pKa = 11.84EE225 pKa = 4.06VLEE228 pKa = 3.84QQRR231 pKa = 11.84DD232 pKa = 3.69LVFNYY237 pKa = 10.37ADD239 pKa = 3.37EE240 pKa = 5.75HH241 pKa = 5.24EE242 pKa = 4.55TQMQLLMSGDD252 pKa = 3.81ASLGTHH258 pKa = 5.42TDD260 pKa = 2.97GRR262 pKa = 11.84AFRR265 pKa = 11.84AMYY268 pKa = 10.49NHH270 pKa = 6.9GADD273 pKa = 3.56VDD275 pKa = 3.7WFIPEE280 pKa = 4.11EE281 pKa = 4.33GTVVGPNEE289 pKa = 3.87VAIPTGGPNPITSTMYY305 pKa = 10.15TDD307 pKa = 4.21YY308 pKa = 11.41LFTDD312 pKa = 3.47EE313 pKa = 5.48GMEE316 pKa = 4.09KK317 pKa = 10.24LVEE320 pKa = 4.28TTVYY324 pKa = 9.89RR325 pKa = 11.84PPMEE329 pKa = 4.86NEE331 pKa = 3.57AFTDD335 pKa = 4.29GEE337 pKa = 4.25FGDD340 pKa = 4.47TIRR343 pKa = 11.84DD344 pKa = 3.27KK345 pKa = 11.04WDD347 pKa = 3.99DD348 pKa = 3.44EE349 pKa = 4.38WEE351 pKa = 4.28KK352 pKa = 11.32EE353 pKa = 3.63GDD355 pKa = 3.64AEE357 pKa = 5.75DD358 pKa = 5.56FIDD361 pKa = 5.32DD362 pKa = 4.26LVITEE367 pKa = 4.85DD368 pKa = 3.65EE369 pKa = 4.5LEE371 pKa = 4.14RR372 pKa = 11.84SYY374 pKa = 11.43DD375 pKa = 2.96AWPRR379 pKa = 11.84SDD381 pKa = 5.59DD382 pKa = 3.7VIEE385 pKa = 5.13RR386 pKa = 11.84YY387 pKa = 10.38DD388 pKa = 4.11EE389 pKa = 3.69IWTEE393 pKa = 3.71ITTGG397 pKa = 3.27
MM1 pKa = 6.2VHH3 pKa = 6.53RR4 pKa = 11.84RR5 pKa = 11.84NVLKK9 pKa = 10.63GAGAISVAGMAGCLGGDD26 pKa = 3.52DD27 pKa = 4.58GEE29 pKa = 4.45YY30 pKa = 9.81TRR32 pKa = 11.84PVEE35 pKa = 4.93IDD37 pKa = 3.27FDD39 pKa = 3.82DD40 pKa = 4.27WPPEE44 pKa = 3.88EE45 pKa = 4.98YY46 pKa = 10.69GDD48 pKa = 5.77SLHH51 pKa = 6.28AWNWYY56 pKa = 9.36GEE58 pKa = 4.15WNEE61 pKa = 3.73WGAEE65 pKa = 4.04NFAEE69 pKa = 4.64EE70 pKa = 4.63YY71 pKa = 10.61DD72 pKa = 3.65LSSYY76 pKa = 7.42EE77 pKa = 4.18TEE79 pKa = 4.58VYY81 pKa = 7.94ATPSDD86 pKa = 3.43WFTNIQANPDD96 pKa = 3.08NHH98 pKa = 7.04GIDD101 pKa = 3.44HH102 pKa = 6.61VGLSTEE108 pKa = 4.04WIHH111 pKa = 7.22RR112 pKa = 11.84VNQEE116 pKa = 4.11DD117 pKa = 3.92ALEE120 pKa = 4.84PIPVDD125 pKa = 3.3EE126 pKa = 5.14LPNVDD131 pKa = 4.61VADD134 pKa = 3.9QYY136 pKa = 11.4MDD138 pKa = 3.0LHH140 pKa = 6.85RR141 pKa = 11.84EE142 pKa = 4.04HH143 pKa = 6.97FGSDD147 pKa = 3.18DD148 pKa = 3.76GVGGVYY154 pKa = 10.23AVPHH158 pKa = 6.89AIVLGPVVVYY168 pKa = 7.81NTNEE172 pKa = 3.77IQDD175 pKa = 4.05PPEE178 pKa = 5.91SIDD181 pKa = 3.47ILWDD185 pKa = 3.13EE186 pKa = 4.79EE187 pKa = 4.4YY188 pKa = 11.2ADD190 pKa = 5.12EE191 pKa = 5.07ISMMAHH197 pKa = 6.99LSGFLCEE204 pKa = 5.45AGALYY209 pKa = 9.84TGQDD213 pKa = 3.8PNDD216 pKa = 4.06PDD218 pKa = 3.78DD219 pKa = 4.35FEE221 pKa = 5.39EE222 pKa = 4.09IRR224 pKa = 11.84EE225 pKa = 4.06VLEE228 pKa = 3.84QQRR231 pKa = 11.84DD232 pKa = 3.69LVFNYY237 pKa = 10.37ADD239 pKa = 3.37EE240 pKa = 5.75HH241 pKa = 5.24EE242 pKa = 4.55TQMQLLMSGDD252 pKa = 3.81ASLGTHH258 pKa = 5.42TDD260 pKa = 2.97GRR262 pKa = 11.84AFRR265 pKa = 11.84AMYY268 pKa = 10.49NHH270 pKa = 6.9GADD273 pKa = 3.56VDD275 pKa = 3.7WFIPEE280 pKa = 4.11EE281 pKa = 4.33GTVVGPNEE289 pKa = 3.87VAIPTGGPNPITSTMYY305 pKa = 10.15TDD307 pKa = 4.21YY308 pKa = 11.41LFTDD312 pKa = 3.47EE313 pKa = 5.48GMEE316 pKa = 4.09KK317 pKa = 10.24LVEE320 pKa = 4.28TTVYY324 pKa = 9.89RR325 pKa = 11.84PPMEE329 pKa = 4.86NEE331 pKa = 3.57AFTDD335 pKa = 4.29GEE337 pKa = 4.25FGDD340 pKa = 4.47TIRR343 pKa = 11.84DD344 pKa = 3.27KK345 pKa = 11.04WDD347 pKa = 3.99DD348 pKa = 3.44EE349 pKa = 4.38WEE351 pKa = 4.28KK352 pKa = 11.32EE353 pKa = 3.63GDD355 pKa = 3.64AEE357 pKa = 5.75DD358 pKa = 5.56FIDD361 pKa = 5.32DD362 pKa = 4.26LVITEE367 pKa = 4.85DD368 pKa = 3.65EE369 pKa = 4.5LEE371 pKa = 4.14RR372 pKa = 11.84SYY374 pKa = 11.43DD375 pKa = 2.96AWPRR379 pKa = 11.84SDD381 pKa = 5.59DD382 pKa = 3.7VIEE385 pKa = 5.13RR386 pKa = 11.84YY387 pKa = 10.38DD388 pKa = 4.11EE389 pKa = 3.69IWTEE393 pKa = 3.71ITTGG397 pKa = 3.27
Molecular weight: 45.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D3SYS5|D3SYS5_NATMM DUF3054 family protein OS=Natrialba magadii (strain ATCC 43099 / DSM 3394 / CCM 3739 / CIP 104546 / IAM 13178 / JCM 8861 / NBRC 102185 / NCIMB 2190 / MS3) OX=547559 GN=Nmag_0600 PE=4 SV=1
MM1 pKa = 7.28ATLTKK6 pKa = 10.14SVPKK10 pKa = 10.38RR11 pKa = 11.84PIGRR15 pKa = 11.84NPRR18 pKa = 11.84VHH20 pKa = 6.63HH21 pKa = 5.87LLGRR25 pKa = 11.84FCLGRR30 pKa = 11.84PEE32 pKa = 4.32GGTRR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84DD40 pKa = 3.62CDD42 pKa = 2.87RR43 pKa = 11.84LSQVRR48 pKa = 11.84GRR50 pKa = 11.84VDD52 pKa = 2.67GWLFGNQSVCDD63 pKa = 3.47RR64 pKa = 11.84VQVQEE69 pKa = 4.23PVPVSEE75 pKa = 4.63VCLKK79 pKa = 9.66TALVCLLTGGRR90 pKa = 11.84NTHH93 pKa = 6.07SARR96 pKa = 11.84DD97 pKa = 3.67PAGVTVFAVCSRR109 pKa = 11.84ADD111 pKa = 3.97GPLAWAAGSTIPRR124 pKa = 11.84SVLSRR129 pKa = 11.84GSHH132 pKa = 5.37SLCSLQTALAPNRR145 pKa = 11.84ARR147 pKa = 11.84CRR149 pKa = 11.84PRR151 pKa = 11.84LLRR154 pKa = 11.84TTRR157 pKa = 11.84SSWDD161 pKa = 3.21GCAVSYY167 pKa = 11.09SDD169 pKa = 5.25YY170 pKa = 11.39YY171 pKa = 11.77SNNCNN176 pKa = 3.41
MM1 pKa = 7.28ATLTKK6 pKa = 10.14SVPKK10 pKa = 10.38RR11 pKa = 11.84PIGRR15 pKa = 11.84NPRR18 pKa = 11.84VHH20 pKa = 6.63HH21 pKa = 5.87LLGRR25 pKa = 11.84FCLGRR30 pKa = 11.84PEE32 pKa = 4.32GGTRR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84DD40 pKa = 3.62CDD42 pKa = 2.87RR43 pKa = 11.84LSQVRR48 pKa = 11.84GRR50 pKa = 11.84VDD52 pKa = 2.67GWLFGNQSVCDD63 pKa = 3.47RR64 pKa = 11.84VQVQEE69 pKa = 4.23PVPVSEE75 pKa = 4.63VCLKK79 pKa = 9.66TALVCLLTGGRR90 pKa = 11.84NTHH93 pKa = 6.07SARR96 pKa = 11.84DD97 pKa = 3.67PAGVTVFAVCSRR109 pKa = 11.84ADD111 pKa = 3.97GPLAWAAGSTIPRR124 pKa = 11.84SVLSRR129 pKa = 11.84GSHH132 pKa = 5.37SLCSLQTALAPNRR145 pKa = 11.84ARR147 pKa = 11.84CRR149 pKa = 11.84PRR151 pKa = 11.84LLRR154 pKa = 11.84TTRR157 pKa = 11.84SSWDD161 pKa = 3.21GCAVSYY167 pKa = 11.09SDD169 pKa = 5.25YY170 pKa = 11.39YY171 pKa = 11.77SNNCNN176 pKa = 3.41
Molecular weight: 19.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1227618 |
30 |
1773 |
292.1 |
31.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.293 ± 0.045 | 0.756 ± 0.012 |
8.786 ± 0.057 | 9.132 ± 0.064 |
3.201 ± 0.025 | 8.14 ± 0.035 |
2.136 ± 0.019 | 4.553 ± 0.033 |
1.774 ± 0.026 | 8.686 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.736 ± 0.016 | 2.502 ± 0.022 |
4.567 ± 0.026 | 2.818 ± 0.021 |
5.952 ± 0.037 | 5.966 ± 0.033 |
6.874 ± 0.034 | 8.314 ± 0.037 |
1.127 ± 0.014 | 2.688 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
