
Capybara microvirus Cap3_SP_391
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.63
Get precalculated fractions of proteins
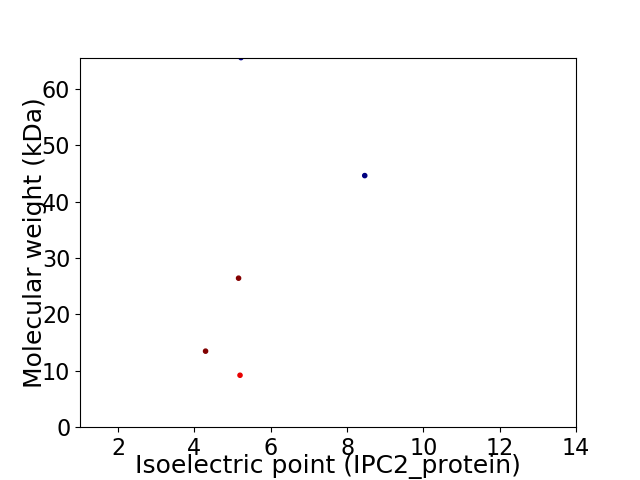
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8H6|A0A4P8W8H6_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_391 OX=2585442 PE=4 SV=1
MM1 pKa = 7.98DD2 pKa = 4.37SEE4 pKa = 4.04EE5 pKa = 5.07DD6 pKa = 3.26IYY8 pKa = 11.61EE9 pKa = 4.46KK10 pKa = 10.55IQADD14 pKa = 3.95LEE16 pKa = 4.27DD17 pKa = 4.2SKK19 pKa = 11.31IEE21 pKa = 4.11NIIQRR26 pKa = 11.84VEE28 pKa = 4.04YY29 pKa = 10.28GDD31 pKa = 3.83VGALSARR38 pKa = 11.84QGIYY42 pKa = 10.69ADD44 pKa = 3.5IADD47 pKa = 5.0MPTSWAEE54 pKa = 3.89AQTKK58 pKa = 7.39ITAMEE63 pKa = 4.1QIFNKK68 pKa = 10.37LPLEE72 pKa = 3.99VRR74 pKa = 11.84KK75 pKa = 10.06AYY77 pKa = 10.45DD78 pKa = 3.26FNFRR82 pKa = 11.84AFIADD87 pKa = 3.62YY88 pKa = 11.05GSDD91 pKa = 3.19QFMEE95 pKa = 4.17ALGVHH100 pKa = 6.43RR101 pKa = 11.84EE102 pKa = 4.18GKK104 pKa = 9.54PVEE107 pKa = 4.1QPAAEE112 pKa = 4.12KK113 pKa = 10.74PEE115 pKa = 4.35GGEE118 pKa = 4.11AEE120 pKa = 4.15
MM1 pKa = 7.98DD2 pKa = 4.37SEE4 pKa = 4.04EE5 pKa = 5.07DD6 pKa = 3.26IYY8 pKa = 11.61EE9 pKa = 4.46KK10 pKa = 10.55IQADD14 pKa = 3.95LEE16 pKa = 4.27DD17 pKa = 4.2SKK19 pKa = 11.31IEE21 pKa = 4.11NIIQRR26 pKa = 11.84VEE28 pKa = 4.04YY29 pKa = 10.28GDD31 pKa = 3.83VGALSARR38 pKa = 11.84QGIYY42 pKa = 10.69ADD44 pKa = 3.5IADD47 pKa = 5.0MPTSWAEE54 pKa = 3.89AQTKK58 pKa = 7.39ITAMEE63 pKa = 4.1QIFNKK68 pKa = 10.37LPLEE72 pKa = 3.99VRR74 pKa = 11.84KK75 pKa = 10.06AYY77 pKa = 10.45DD78 pKa = 3.26FNFRR82 pKa = 11.84AFIADD87 pKa = 3.62YY88 pKa = 11.05GSDD91 pKa = 3.19QFMEE95 pKa = 4.17ALGVHH100 pKa = 6.43RR101 pKa = 11.84EE102 pKa = 4.18GKK104 pKa = 9.54PVEE107 pKa = 4.1QPAAEE112 pKa = 4.12KK113 pKa = 10.74PEE115 pKa = 4.35GGEE118 pKa = 4.11AEE120 pKa = 4.15
Molecular weight: 13.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4W5|A0A4P8W4W5_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_391 OX=2585442 PE=3 SV=1
MM1 pKa = 7.71AWRR4 pKa = 11.84AYY6 pKa = 9.63CIAWQPITGCSRR18 pKa = 11.84NCSKK22 pKa = 10.52IAAGTRR28 pKa = 11.84RR29 pKa = 11.84AYY31 pKa = 10.28RR32 pKa = 11.84KK33 pKa = 9.3QRR35 pKa = 11.84WGLMRR40 pKa = 11.84CSHH43 pKa = 7.28PLMAFYY49 pKa = 10.09TIDD52 pKa = 3.54PVTGRR57 pKa = 11.84KK58 pKa = 8.35NLRR61 pKa = 11.84HH62 pKa = 6.03FKK64 pKa = 10.84SIDD67 pKa = 3.42TADD70 pKa = 3.14GKK72 pKa = 10.76AIYY75 pKa = 9.01EE76 pKa = 4.05LHH78 pKa = 6.03DD79 pKa = 3.49QEE81 pKa = 4.34RR82 pKa = 11.84TIMIPCGQCLACRR95 pKa = 11.84MRR97 pKa = 11.84YY98 pKa = 9.78SNDD101 pKa = 2.75WANRR105 pKa = 11.84IYY107 pKa = 10.98LEE109 pKa = 4.22ACTYY113 pKa = 10.31PRR115 pKa = 11.84DD116 pKa = 4.08RR117 pKa = 11.84NWFLTLTYY125 pKa = 10.84DD126 pKa = 4.76EE127 pKa = 5.18DD128 pKa = 3.85HH129 pKa = 6.82LPYY132 pKa = 10.78GSGEE136 pKa = 4.16RR137 pKa = 11.84PTLIKK142 pKa = 10.93DD143 pKa = 3.85EE144 pKa = 4.2ISKK147 pKa = 10.17FMKK150 pKa = 10.38ALRR153 pKa = 11.84QRR155 pKa = 11.84WAEE158 pKa = 4.4DD159 pKa = 3.2EE160 pKa = 4.36TQTEE164 pKa = 5.01GIRR167 pKa = 11.84FFGCGEE173 pKa = 4.15YY174 pKa = 10.58GGKK177 pKa = 6.81TQRR180 pKa = 11.84PHH182 pKa = 4.92YY183 pKa = 10.26HH184 pKa = 7.05IILLNCLIKK193 pKa = 10.74KK194 pKa = 10.03EE195 pKa = 3.95DD196 pKa = 3.48LKK198 pKa = 11.34VYY200 pKa = 8.84KK201 pKa = 10.28TEE203 pKa = 4.44AGHH206 pKa = 6.4VLWNVPKK213 pKa = 9.46ITHH216 pKa = 5.64IWGKK220 pKa = 8.07GHH222 pKa = 6.5VVVAEE227 pKa = 4.65CNWEE231 pKa = 4.03TCAYY235 pKa = 7.74TSRR238 pKa = 11.84YY239 pKa = 7.47MLKK242 pKa = 9.72KK243 pKa = 10.14HH244 pKa = 6.26KK245 pKa = 10.68GQDD248 pKa = 2.65ADD250 pKa = 3.78YY251 pKa = 9.76YY252 pKa = 9.18EE253 pKa = 4.54KK254 pKa = 11.04LGIYY258 pKa = 9.31PEE260 pKa = 4.07HH261 pKa = 6.25TRR263 pKa = 11.84MSRR266 pKa = 11.84RR267 pKa = 11.84PGIGADD273 pKa = 3.51YY274 pKa = 11.14LDD276 pKa = 5.61DD277 pKa = 3.45NWEE280 pKa = 4.33QIWSRR285 pKa = 11.84DD286 pKa = 3.32HH287 pKa = 7.87ILIKK291 pKa = 10.29GHH293 pKa = 5.14QAAVPAAYY301 pKa = 8.42TRR303 pKa = 11.84KK304 pKa = 10.08LGYY307 pKa = 9.99SDD309 pKa = 5.1LDD311 pKa = 3.48RR312 pKa = 11.84EE313 pKa = 4.56AEE315 pKa = 4.0IKK317 pKa = 9.96AKK319 pKa = 10.24RR320 pKa = 11.84MHH322 pKa = 6.83RR323 pKa = 11.84AQLTAEE329 pKa = 4.03NEE331 pKa = 4.36ANLGAYY337 pKa = 7.9DD338 pKa = 3.81RR339 pKa = 11.84PVWEE343 pKa = 3.81QLQIEE348 pKa = 4.57EE349 pKa = 3.93EE350 pKa = 4.16AMRR353 pKa = 11.84QRR355 pKa = 11.84TALLKK360 pKa = 10.43RR361 pKa = 11.84RR362 pKa = 11.84LEE364 pKa = 3.98KK365 pKa = 10.96GILKK369 pKa = 10.11KK370 pKa = 10.42KK371 pKa = 7.55YY372 pKa = 9.26HH373 pKa = 6.49LYY375 pKa = 10.36EE376 pKa = 4.69GSLII380 pKa = 3.97
MM1 pKa = 7.71AWRR4 pKa = 11.84AYY6 pKa = 9.63CIAWQPITGCSRR18 pKa = 11.84NCSKK22 pKa = 10.52IAAGTRR28 pKa = 11.84RR29 pKa = 11.84AYY31 pKa = 10.28RR32 pKa = 11.84KK33 pKa = 9.3QRR35 pKa = 11.84WGLMRR40 pKa = 11.84CSHH43 pKa = 7.28PLMAFYY49 pKa = 10.09TIDD52 pKa = 3.54PVTGRR57 pKa = 11.84KK58 pKa = 8.35NLRR61 pKa = 11.84HH62 pKa = 6.03FKK64 pKa = 10.84SIDD67 pKa = 3.42TADD70 pKa = 3.14GKK72 pKa = 10.76AIYY75 pKa = 9.01EE76 pKa = 4.05LHH78 pKa = 6.03DD79 pKa = 3.49QEE81 pKa = 4.34RR82 pKa = 11.84TIMIPCGQCLACRR95 pKa = 11.84MRR97 pKa = 11.84YY98 pKa = 9.78SNDD101 pKa = 2.75WANRR105 pKa = 11.84IYY107 pKa = 10.98LEE109 pKa = 4.22ACTYY113 pKa = 10.31PRR115 pKa = 11.84DD116 pKa = 4.08RR117 pKa = 11.84NWFLTLTYY125 pKa = 10.84DD126 pKa = 4.76EE127 pKa = 5.18DD128 pKa = 3.85HH129 pKa = 6.82LPYY132 pKa = 10.78GSGEE136 pKa = 4.16RR137 pKa = 11.84PTLIKK142 pKa = 10.93DD143 pKa = 3.85EE144 pKa = 4.2ISKK147 pKa = 10.17FMKK150 pKa = 10.38ALRR153 pKa = 11.84QRR155 pKa = 11.84WAEE158 pKa = 4.4DD159 pKa = 3.2EE160 pKa = 4.36TQTEE164 pKa = 5.01GIRR167 pKa = 11.84FFGCGEE173 pKa = 4.15YY174 pKa = 10.58GGKK177 pKa = 6.81TQRR180 pKa = 11.84PHH182 pKa = 4.92YY183 pKa = 10.26HH184 pKa = 7.05IILLNCLIKK193 pKa = 10.74KK194 pKa = 10.03EE195 pKa = 3.95DD196 pKa = 3.48LKK198 pKa = 11.34VYY200 pKa = 8.84KK201 pKa = 10.28TEE203 pKa = 4.44AGHH206 pKa = 6.4VLWNVPKK213 pKa = 9.46ITHH216 pKa = 5.64IWGKK220 pKa = 8.07GHH222 pKa = 6.5VVVAEE227 pKa = 4.65CNWEE231 pKa = 4.03TCAYY235 pKa = 7.74TSRR238 pKa = 11.84YY239 pKa = 7.47MLKK242 pKa = 9.72KK243 pKa = 10.14HH244 pKa = 6.26KK245 pKa = 10.68GQDD248 pKa = 2.65ADD250 pKa = 3.78YY251 pKa = 9.76YY252 pKa = 9.18EE253 pKa = 4.54KK254 pKa = 11.04LGIYY258 pKa = 9.31PEE260 pKa = 4.07HH261 pKa = 6.25TRR263 pKa = 11.84MSRR266 pKa = 11.84RR267 pKa = 11.84PGIGADD273 pKa = 3.51YY274 pKa = 11.14LDD276 pKa = 5.61DD277 pKa = 3.45NWEE280 pKa = 4.33QIWSRR285 pKa = 11.84DD286 pKa = 3.32HH287 pKa = 7.87ILIKK291 pKa = 10.29GHH293 pKa = 5.14QAAVPAAYY301 pKa = 8.42TRR303 pKa = 11.84KK304 pKa = 10.08LGYY307 pKa = 9.99SDD309 pKa = 5.1LDD311 pKa = 3.48RR312 pKa = 11.84EE313 pKa = 4.56AEE315 pKa = 4.0IKK317 pKa = 9.96AKK319 pKa = 10.24RR320 pKa = 11.84MHH322 pKa = 6.83RR323 pKa = 11.84AQLTAEE329 pKa = 4.03NEE331 pKa = 4.36ANLGAYY337 pKa = 7.9DD338 pKa = 3.81RR339 pKa = 11.84PVWEE343 pKa = 3.81QLQIEE348 pKa = 4.57EE349 pKa = 3.93EE350 pKa = 4.16AMRR353 pKa = 11.84QRR355 pKa = 11.84TALLKK360 pKa = 10.43RR361 pKa = 11.84RR362 pKa = 11.84LEE364 pKa = 3.98KK365 pKa = 10.96GILKK369 pKa = 10.11KK370 pKa = 10.42KK371 pKa = 7.55YY372 pKa = 9.26HH373 pKa = 6.49LYY375 pKa = 10.36EE376 pKa = 4.69GSLII380 pKa = 3.97
Molecular weight: 44.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1413 |
82 |
579 |
282.6 |
31.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.13 ± 1.16 | 1.132 ± 0.658 |
5.591 ± 0.313 | 6.369 ± 1.064 |
3.043 ± 0.542 | 7.573 ± 1.065 |
1.84 ± 0.762 | 5.662 ± 0.711 |
5.308 ± 0.739 | 6.369 ± 0.662 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.609 ± 0.548 | 4.105 ± 0.588 |
4.388 ± 0.8 | 5.096 ± 0.455 |
5.449 ± 1.206 | 6.794 ± 1.762 |
6.369 ± 1.074 | 4.388 ± 0.779 |
2.265 ± 0.514 | 5.52 ± 0.686 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
