
Sphingomonas sp. URHD0007
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
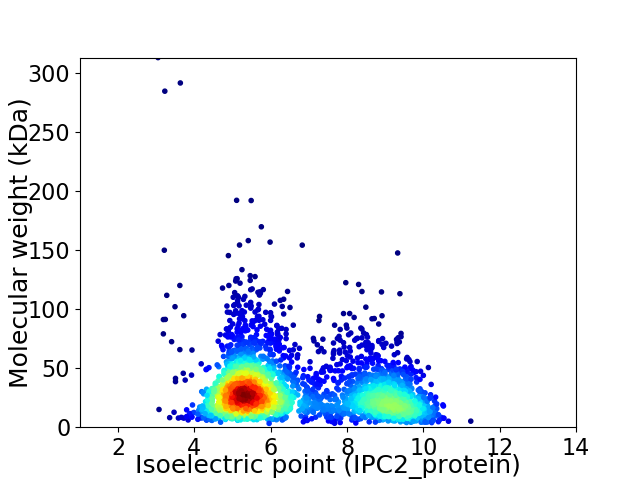
Virtual 2D-PAGE plot for 2847 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U1G197|A0A2U1G197_9SPHN Non-homologous end joining protein Ku OS=Sphingomonas sp. URHD0007 OX=1298856 GN=ku PE=3 SV=1
MM1 pKa = 7.65PRR3 pKa = 11.84TTRR6 pKa = 11.84GPIDD10 pKa = 4.1LLSGYY15 pKa = 8.16TIIDD19 pKa = 3.76PVVGDD24 pKa = 4.08PAEE27 pKa = 3.73QDD29 pKa = 3.67YY30 pKa = 11.51YY31 pKa = 11.6EE32 pKa = 4.78MGNLTIWGVGLITFGPPTAAQQAWVADD59 pKa = 3.71GANYY63 pKa = 10.67SNLATFPGDD72 pKa = 2.89WVSFGFPIYY81 pKa = 10.28FPNATYY87 pKa = 10.57YY88 pKa = 8.19ATPIPIWSTAHH99 pKa = 5.76MVDD102 pKa = 3.34GVIVNEE108 pKa = 4.03QGIKK112 pKa = 10.52GADD115 pKa = 2.84AWGIAGQTGTGTYY128 pKa = 9.49FFSDD132 pKa = 3.14ALVTNGTNASEE143 pKa = 4.37TLTGTASAEE152 pKa = 4.22TLNGLGGDD160 pKa = 3.67DD161 pKa = 4.08TLIGGAGSDD170 pKa = 3.95ALHH173 pKa = 6.56GGAGNDD179 pKa = 3.79HH180 pKa = 6.67LVGGAGVDD188 pKa = 4.0RR189 pKa = 11.84LWGDD193 pKa = 3.82DD194 pKa = 3.88GNDD197 pKa = 3.48VLDD200 pKa = 5.04PGSDD204 pKa = 2.9AAFIYY209 pKa = 10.09VASDD213 pKa = 3.06VNRR216 pKa = 11.84AGVYY220 pKa = 9.96FVDD223 pKa = 4.86GGAGLDD229 pKa = 3.79TLVLDD234 pKa = 4.14YY235 pKa = 11.29SAATRR240 pKa = 11.84SQSISGDD247 pKa = 3.41QVLASPQVVNVEE259 pKa = 4.11ALQITGSAFSDD270 pKa = 3.83FLSGSSNADD279 pKa = 3.11QLYY282 pKa = 10.21GGGGFDD288 pKa = 3.9HH289 pKa = 7.3LSGGGGNDD297 pKa = 3.95LLDD300 pKa = 4.37AGAPGSSSVTTIGAGGHH317 pKa = 4.96STADD321 pKa = 3.54ALSLDD326 pKa = 3.69HH327 pKa = 6.91LFNAGATAPTVTFSITQTEE346 pKa = 4.57VNVVNWGFRR355 pKa = 11.84PVAGTVYY362 pKa = 10.69SFTVADD368 pKa = 4.45ASDD371 pKa = 3.54QAWIDD376 pKa = 3.48FSLTDD381 pKa = 3.98FGSEE385 pKa = 3.83VLYY388 pKa = 10.94GFTITDD394 pKa = 3.44SDD396 pKa = 4.14GVAVDD401 pKa = 3.49WFPYY405 pKa = 10.6DD406 pKa = 3.37PTVPIAFPHH415 pKa = 6.44AGTYY419 pKa = 10.09FLTVDD424 pKa = 3.71VTNTNPWAEE433 pKa = 3.98AGIDD437 pKa = 3.52VTLTLEE443 pKa = 4.51GADD446 pKa = 3.26VLAANVLEE454 pKa = 5.09GGTGDD459 pKa = 3.59DD460 pKa = 3.77TYY462 pKa = 11.79VVYY465 pKa = 10.69SATDD469 pKa = 3.27QVVEE473 pKa = 4.21NPGEE477 pKa = 4.2GTDD480 pKa = 3.7TVRR483 pKa = 11.84SSISFALVANVEE495 pKa = 4.07NLTLTGAAAINGTGNGLANIITGNAAANVITGGGGGDD532 pKa = 3.79TLTGGAGADD541 pKa = 3.4TFRR544 pKa = 11.84YY545 pKa = 9.18VALSDD550 pKa = 3.91STSAAADD557 pKa = 5.07LITDD561 pKa = 4.41FIGGKK566 pKa = 9.95KK567 pKa = 10.47GGDD570 pKa = 3.81KK571 pKa = 10.17IDD573 pKa = 4.56LSAIDD578 pKa = 4.68ANINTLANDD587 pKa = 3.7AFHH590 pKa = 6.65LVKK593 pKa = 10.74GFTGHH598 pKa = 7.32AGEE601 pKa = 5.79LYY603 pKa = 10.21TSYY606 pKa = 11.34DD607 pKa = 3.34KK608 pKa = 11.62AGGTTNIYY616 pKa = 10.67LDD618 pKa = 3.57VDD620 pKa = 3.82GDD622 pKa = 3.79RR623 pKa = 11.84AADD626 pKa = 3.71MVIHH630 pKa = 6.13LTGHH634 pKa = 6.95LNLASGDD641 pKa = 4.04FIFF644 pKa = 5.77
MM1 pKa = 7.65PRR3 pKa = 11.84TTRR6 pKa = 11.84GPIDD10 pKa = 4.1LLSGYY15 pKa = 8.16TIIDD19 pKa = 3.76PVVGDD24 pKa = 4.08PAEE27 pKa = 3.73QDD29 pKa = 3.67YY30 pKa = 11.51YY31 pKa = 11.6EE32 pKa = 4.78MGNLTIWGVGLITFGPPTAAQQAWVADD59 pKa = 3.71GANYY63 pKa = 10.67SNLATFPGDD72 pKa = 2.89WVSFGFPIYY81 pKa = 10.28FPNATYY87 pKa = 10.57YY88 pKa = 8.19ATPIPIWSTAHH99 pKa = 5.76MVDD102 pKa = 3.34GVIVNEE108 pKa = 4.03QGIKK112 pKa = 10.52GADD115 pKa = 2.84AWGIAGQTGTGTYY128 pKa = 9.49FFSDD132 pKa = 3.14ALVTNGTNASEE143 pKa = 4.37TLTGTASAEE152 pKa = 4.22TLNGLGGDD160 pKa = 3.67DD161 pKa = 4.08TLIGGAGSDD170 pKa = 3.95ALHH173 pKa = 6.56GGAGNDD179 pKa = 3.79HH180 pKa = 6.67LVGGAGVDD188 pKa = 4.0RR189 pKa = 11.84LWGDD193 pKa = 3.82DD194 pKa = 3.88GNDD197 pKa = 3.48VLDD200 pKa = 5.04PGSDD204 pKa = 2.9AAFIYY209 pKa = 10.09VASDD213 pKa = 3.06VNRR216 pKa = 11.84AGVYY220 pKa = 9.96FVDD223 pKa = 4.86GGAGLDD229 pKa = 3.79TLVLDD234 pKa = 4.14YY235 pKa = 11.29SAATRR240 pKa = 11.84SQSISGDD247 pKa = 3.41QVLASPQVVNVEE259 pKa = 4.11ALQITGSAFSDD270 pKa = 3.83FLSGSSNADD279 pKa = 3.11QLYY282 pKa = 10.21GGGGFDD288 pKa = 3.9HH289 pKa = 7.3LSGGGGNDD297 pKa = 3.95LLDD300 pKa = 4.37AGAPGSSSVTTIGAGGHH317 pKa = 4.96STADD321 pKa = 3.54ALSLDD326 pKa = 3.69HH327 pKa = 6.91LFNAGATAPTVTFSITQTEE346 pKa = 4.57VNVVNWGFRR355 pKa = 11.84PVAGTVYY362 pKa = 10.69SFTVADD368 pKa = 4.45ASDD371 pKa = 3.54QAWIDD376 pKa = 3.48FSLTDD381 pKa = 3.98FGSEE385 pKa = 3.83VLYY388 pKa = 10.94GFTITDD394 pKa = 3.44SDD396 pKa = 4.14GVAVDD401 pKa = 3.49WFPYY405 pKa = 10.6DD406 pKa = 3.37PTVPIAFPHH415 pKa = 6.44AGTYY419 pKa = 10.09FLTVDD424 pKa = 3.71VTNTNPWAEE433 pKa = 3.98AGIDD437 pKa = 3.52VTLTLEE443 pKa = 4.51GADD446 pKa = 3.26VLAANVLEE454 pKa = 5.09GGTGDD459 pKa = 3.59DD460 pKa = 3.77TYY462 pKa = 11.79VVYY465 pKa = 10.69SATDD469 pKa = 3.27QVVEE473 pKa = 4.21NPGEE477 pKa = 4.2GTDD480 pKa = 3.7TVRR483 pKa = 11.84SSISFALVANVEE495 pKa = 4.07NLTLTGAAAINGTGNGLANIITGNAAANVITGGGGGDD532 pKa = 3.79TLTGGAGADD541 pKa = 3.4TFRR544 pKa = 11.84YY545 pKa = 9.18VALSDD550 pKa = 3.91STSAAADD557 pKa = 5.07LITDD561 pKa = 4.41FIGGKK566 pKa = 9.95KK567 pKa = 10.47GGDD570 pKa = 3.81KK571 pKa = 10.17IDD573 pKa = 4.56LSAIDD578 pKa = 4.68ANINTLANDD587 pKa = 3.7AFHH590 pKa = 6.65LVKK593 pKa = 10.74GFTGHH598 pKa = 7.32AGEE601 pKa = 5.79LYY603 pKa = 10.21TSYY606 pKa = 11.34DD607 pKa = 3.34KK608 pKa = 11.62AGGTTNIYY616 pKa = 10.67LDD618 pKa = 3.57VDD620 pKa = 3.82GDD622 pKa = 3.79RR623 pKa = 11.84AADD626 pKa = 3.71MVIHH630 pKa = 6.13LTGHH634 pKa = 6.95LNLASGDD641 pKa = 4.04FIFF644 pKa = 5.77
Molecular weight: 65.73 kDa
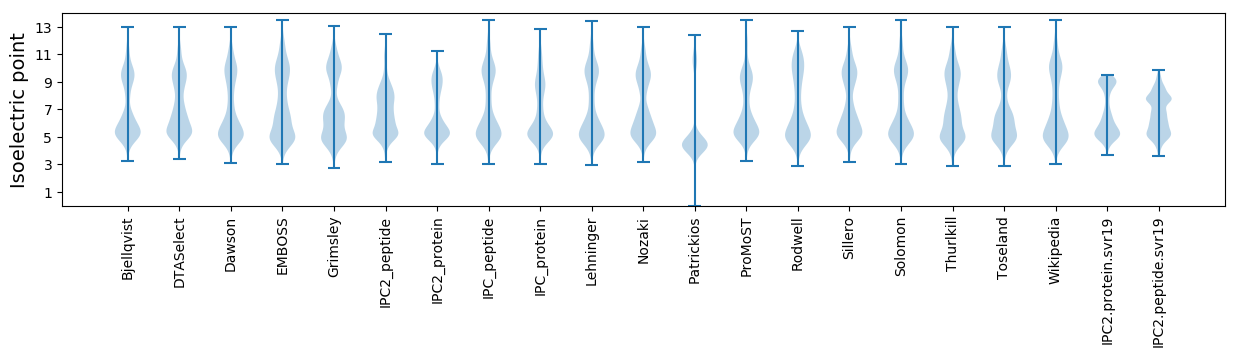
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U1FXB6|A0A2U1FXB6_9SPHN Pili/flagellar assembly PapD-like chaperone OS=Sphingomonas sp. URHD0007 OX=1298856 GN=H952_0249 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.06VLNARR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.06KK41 pKa = 10.57LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.06VLNARR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.06KK41 pKa = 10.57LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
897370 |
29 |
3155 |
315.2 |
34.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.79 ± 0.069 | 0.774 ± 0.016 |
5.926 ± 0.054 | 5.692 ± 0.058 |
3.599 ± 0.026 | 8.83 ± 0.071 |
2.032 ± 0.022 | 4.971 ± 0.032 |
3.151 ± 0.034 | 9.903 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.185 ± 0.021 | 2.643 ± 0.04 |
5.414 ± 0.049 | 3.134 ± 0.026 |
7.329 ± 0.068 | 5.503 ± 0.039 |
5.154 ± 0.05 | 7.271 ± 0.038 |
1.472 ± 0.023 | 2.228 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
