
Actinoplanes missouriensis (strain ATCC 14538 / DSM 43046 / CBS 188.64 / JCM 3121 / NCIMB 12654 / NBRC 102363 / 431)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micromonosporales; Micromonosporaceae; Actinoplanes; Actinoplanes missouriensis
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 8113 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
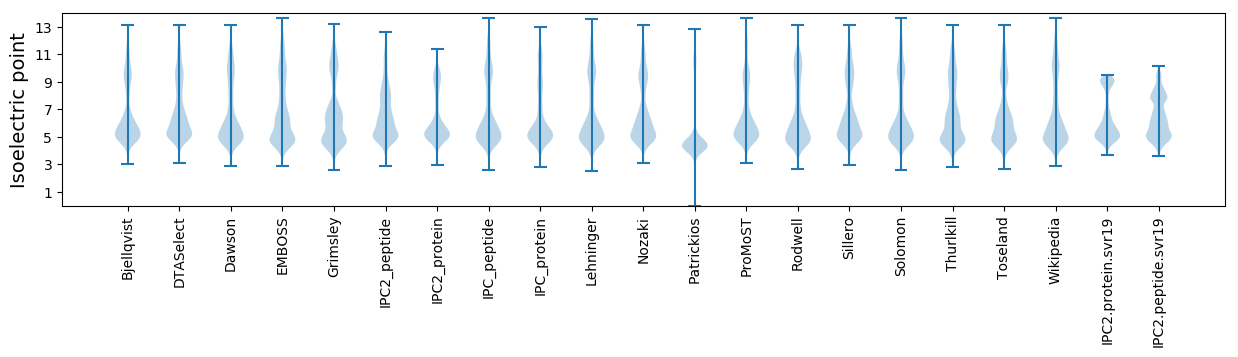
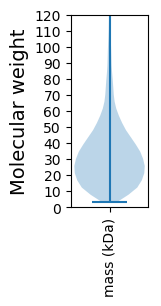
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|I0H5B9|I0H5B9_ACTM4 HintN domain-containing protein OS=Actinoplanes missouriensis (strain ATCC 14538 / DSM 43046 / CBS 188.64 / JCM 3121 / NCIMB 12654 / NBRC 102363 / 431) OX=512565 GN=AMIS_29860 PE=4 SV=1
MM1 pKa = 7.26NVYY4 pKa = 10.3DD5 pKa = 3.47IAGRR9 pKa = 11.84LPSIPVLRR17 pKa = 11.84DD18 pKa = 2.87RR19 pKa = 11.84CRR21 pKa = 11.84ALAVLEE27 pKa = 4.91LIQGTGYY34 pKa = 10.9EE35 pKa = 4.5YY36 pKa = 11.2YY37 pKa = 10.62SYY39 pKa = 9.63TAWGDD44 pKa = 3.55SEE46 pKa = 4.44AALMSNGSGDD56 pKa = 3.43EE57 pKa = 4.01YY58 pKa = 10.97TIVFGDD64 pKa = 3.19AGVFIRR70 pKa = 11.84LFDD73 pKa = 4.14HH74 pKa = 6.8EE75 pKa = 4.99SAMSPYY81 pKa = 10.84AADD84 pKa = 4.78DD85 pKa = 5.99DD86 pKa = 4.19MALWPGLLDD95 pKa = 3.76GLPPEE100 pKa = 4.8FAGQVADD107 pKa = 4.52PAFCDD112 pKa = 3.91GDD114 pKa = 4.44DD115 pKa = 5.04LIATAVLWRR124 pKa = 11.84RR125 pKa = 11.84TGDD128 pKa = 3.82DD129 pKa = 3.0RR130 pKa = 11.84WHH132 pKa = 6.85AGAGITFPPARR143 pKa = 11.84GPYY146 pKa = 9.8DD147 pKa = 3.56ISPDD151 pKa = 3.31GTDD154 pKa = 3.67RR155 pKa = 11.84LSILCDD161 pKa = 4.78DD162 pKa = 4.23IVDD165 pKa = 4.34AYY167 pKa = 11.76VDD169 pKa = 4.25FASDD173 pKa = 3.93YY174 pKa = 11.04YY175 pKa = 10.11EE176 pKa = 4.1TEE178 pKa = 3.59IDD180 pKa = 3.71RR181 pKa = 11.84DD182 pKa = 3.48AVAYY186 pKa = 9.62VAALRR191 pKa = 11.84PLTDD195 pKa = 3.42DD196 pKa = 3.59VVRR199 pKa = 11.84ALNPEE204 pKa = 3.89SSLAAVRR211 pKa = 11.84TEE213 pKa = 4.06VAAAGYY219 pKa = 8.62PIATAVSS226 pKa = 3.52
MM1 pKa = 7.26NVYY4 pKa = 10.3DD5 pKa = 3.47IAGRR9 pKa = 11.84LPSIPVLRR17 pKa = 11.84DD18 pKa = 2.87RR19 pKa = 11.84CRR21 pKa = 11.84ALAVLEE27 pKa = 4.91LIQGTGYY34 pKa = 10.9EE35 pKa = 4.5YY36 pKa = 11.2YY37 pKa = 10.62SYY39 pKa = 9.63TAWGDD44 pKa = 3.55SEE46 pKa = 4.44AALMSNGSGDD56 pKa = 3.43EE57 pKa = 4.01YY58 pKa = 10.97TIVFGDD64 pKa = 3.19AGVFIRR70 pKa = 11.84LFDD73 pKa = 4.14HH74 pKa = 6.8EE75 pKa = 4.99SAMSPYY81 pKa = 10.84AADD84 pKa = 4.78DD85 pKa = 5.99DD86 pKa = 4.19MALWPGLLDD95 pKa = 3.76GLPPEE100 pKa = 4.8FAGQVADD107 pKa = 4.52PAFCDD112 pKa = 3.91GDD114 pKa = 4.44DD115 pKa = 5.04LIATAVLWRR124 pKa = 11.84RR125 pKa = 11.84TGDD128 pKa = 3.82DD129 pKa = 3.0RR130 pKa = 11.84WHH132 pKa = 6.85AGAGITFPPARR143 pKa = 11.84GPYY146 pKa = 9.8DD147 pKa = 3.56ISPDD151 pKa = 3.31GTDD154 pKa = 3.67RR155 pKa = 11.84LSILCDD161 pKa = 4.78DD162 pKa = 4.23IVDD165 pKa = 4.34AYY167 pKa = 11.76VDD169 pKa = 4.25FASDD173 pKa = 3.93YY174 pKa = 11.04YY175 pKa = 10.11EE176 pKa = 4.1TEE178 pKa = 3.59IDD180 pKa = 3.71RR181 pKa = 11.84DD182 pKa = 3.48AVAYY186 pKa = 9.62VAALRR191 pKa = 11.84PLTDD195 pKa = 3.42DD196 pKa = 3.59VVRR199 pKa = 11.84ALNPEE204 pKa = 3.89SSLAAVRR211 pKa = 11.84TEE213 pKa = 4.06VAAAGYY219 pKa = 8.62PIATAVSS226 pKa = 3.52
Molecular weight: 24.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0HE95|I0HE95_ACTM4 Putative alcohol dehydrogenase OS=Actinoplanes missouriensis (strain ATCC 14538 / DSM 43046 / CBS 188.64 / JCM 3121 / NCIMB 12654 / NBRC 102363 / 431) OX=512565 GN=AMIS_61120 PE=3 SV=1
MM1 pKa = 7.14VATAIAVVTRR11 pKa = 11.84VAATGAGSVAVTVTVVVTRR30 pKa = 11.84VAATGAVSVVGTVRR44 pKa = 11.84RR45 pKa = 11.84ATVVVSVVTVRR56 pKa = 11.84RR57 pKa = 11.84VTVRR61 pKa = 11.84RR62 pKa = 11.84VAASVVATVIVAATRR77 pKa = 11.84VAATAVVSVVATVIVAVTRR96 pKa = 11.84VAATAVVSVVATVTGAAIRR115 pKa = 11.84VAATGVVSVVTVRR128 pKa = 11.84RR129 pKa = 11.84GAASVVATVTVAVTRR144 pKa = 11.84VAATAVVSVVATVTVVVSRR163 pKa = 11.84VAATAVVSVVGTVRR177 pKa = 11.84RR178 pKa = 11.84ATAVVSVVTVRR189 pKa = 11.84RR190 pKa = 11.84VTVRR194 pKa = 11.84RR195 pKa = 11.84VAASVAEE202 pKa = 4.29TVTVAVTRR210 pKa = 11.84VAATAVVSVVGTVRR224 pKa = 11.84RR225 pKa = 11.84ATAVVSVVTVRR236 pKa = 11.84RR237 pKa = 11.84VTVRR241 pKa = 11.84RR242 pKa = 11.84VAASVAEE249 pKa = 4.07TAIAVATRR257 pKa = 11.84VAATAAVSAVTGRR270 pKa = 11.84RR271 pKa = 11.84VTGAATRR278 pKa = 11.84VAATAAGSVAGTVTVAVTRR297 pKa = 11.84VAATAAASVAGTVRR311 pKa = 11.84VVRR314 pKa = 11.84VRR316 pKa = 11.84ASRR319 pKa = 11.84ATAAGSVVATVRR331 pKa = 11.84RR332 pKa = 11.84VTVRR336 pKa = 11.84RR337 pKa = 11.84AAASVAEE344 pKa = 4.33TATAVATRR352 pKa = 11.84VAATAAASVVTVRR365 pKa = 11.84RR366 pKa = 11.84VTVRR370 pKa = 11.84RR371 pKa = 11.84VAASVVATATAVATRR386 pKa = 11.84VAATGAGSVVATVTGAATRR405 pKa = 11.84VAATAGATRR414 pKa = 11.84VATVVVTRR422 pKa = 11.84VAATGAGSATTTAVATGGTSGTAPVRR448 pKa = 11.84VRR450 pKa = 11.84RR451 pKa = 11.84TRR453 pKa = 11.84PARR456 pKa = 11.84RR457 pKa = 11.84RR458 pKa = 11.84SSPRR462 pKa = 11.84TFPRR466 pKa = 11.84TSSRR470 pKa = 11.84PTSTRR475 pKa = 11.84RR476 pKa = 11.84SARR479 pKa = 11.84NN480 pKa = 3.02
MM1 pKa = 7.14VATAIAVVTRR11 pKa = 11.84VAATGAGSVAVTVTVVVTRR30 pKa = 11.84VAATGAVSVVGTVRR44 pKa = 11.84RR45 pKa = 11.84ATVVVSVVTVRR56 pKa = 11.84RR57 pKa = 11.84VTVRR61 pKa = 11.84RR62 pKa = 11.84VAASVVATVIVAATRR77 pKa = 11.84VAATAVVSVVATVIVAVTRR96 pKa = 11.84VAATAVVSVVATVTGAAIRR115 pKa = 11.84VAATGVVSVVTVRR128 pKa = 11.84RR129 pKa = 11.84GAASVVATVTVAVTRR144 pKa = 11.84VAATAVVSVVATVTVVVSRR163 pKa = 11.84VAATAVVSVVGTVRR177 pKa = 11.84RR178 pKa = 11.84ATAVVSVVTVRR189 pKa = 11.84RR190 pKa = 11.84VTVRR194 pKa = 11.84RR195 pKa = 11.84VAASVAEE202 pKa = 4.29TVTVAVTRR210 pKa = 11.84VAATAVVSVVGTVRR224 pKa = 11.84RR225 pKa = 11.84ATAVVSVVTVRR236 pKa = 11.84RR237 pKa = 11.84VTVRR241 pKa = 11.84RR242 pKa = 11.84VAASVAEE249 pKa = 4.07TAIAVATRR257 pKa = 11.84VAATAAVSAVTGRR270 pKa = 11.84RR271 pKa = 11.84VTGAATRR278 pKa = 11.84VAATAAGSVAGTVTVAVTRR297 pKa = 11.84VAATAAASVAGTVRR311 pKa = 11.84VVRR314 pKa = 11.84VRR316 pKa = 11.84ASRR319 pKa = 11.84ATAAGSVVATVRR331 pKa = 11.84RR332 pKa = 11.84VTVRR336 pKa = 11.84RR337 pKa = 11.84AAASVAEE344 pKa = 4.33TATAVATRR352 pKa = 11.84VAATAAASVVTVRR365 pKa = 11.84RR366 pKa = 11.84VTVRR370 pKa = 11.84RR371 pKa = 11.84VAASVVATATAVATRR386 pKa = 11.84VAATGAGSVVATVTGAATRR405 pKa = 11.84VAATAGATRR414 pKa = 11.84VATVVVTRR422 pKa = 11.84VAATGAGSATTTAVATGGTSGTAPVRR448 pKa = 11.84VRR450 pKa = 11.84RR451 pKa = 11.84TRR453 pKa = 11.84PARR456 pKa = 11.84RR457 pKa = 11.84RR458 pKa = 11.84SSPRR462 pKa = 11.84TFPRR466 pKa = 11.84TSSRR470 pKa = 11.84PTSTRR475 pKa = 11.84RR476 pKa = 11.84SARR479 pKa = 11.84NN480 pKa = 3.02
Molecular weight: 46.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2638011 |
30 |
4154 |
325.2 |
34.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.932 ± 0.047 | 0.692 ± 0.007 |
5.96 ± 0.021 | 5.234 ± 0.028 |
2.789 ± 0.017 | 9.26 ± 0.028 |
2.081 ± 0.013 | 3.744 ± 0.017 |
1.953 ± 0.021 | 10.312 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.775 ± 0.009 | 1.93 ± 0.016 |
6.012 ± 0.029 | 2.768 ± 0.014 |
7.894 ± 0.03 | 5.15 ± 0.021 |
6.242 ± 0.027 | 8.65 ± 0.023 |
1.574 ± 0.013 | 2.048 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
