
Sophora alopecuroides yellow stunt alphasatellite 8
Taxonomy: Viruses; Alphasatellitidae; Nanoalphasatellitinae; Clostunsatellite; Sophora yellow stunt alphasatellite 5
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W5YSH3|A0A1W5YSH3_9VIRU Satellite replication initiator protein OS=Sophora alopecuroides yellow stunt alphasatellite 8 OX=1980173 PE=4 SV=1
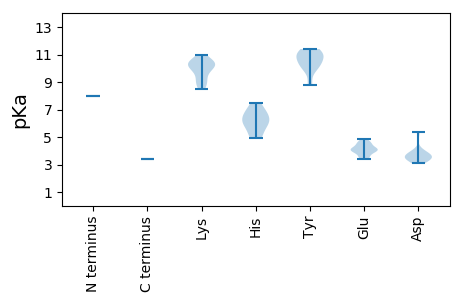
MM1 pKa = 7.98PSQKK5 pKa = 8.99STCWVFTLNFEE16 pKa = 4.51GDD18 pKa = 3.83PPTLTFDD25 pKa = 5.33DD26 pKa = 4.08RR27 pKa = 11.84VQYY30 pKa = 11.05ASWQHH35 pKa = 5.1EE36 pKa = 4.64RR37 pKa = 11.84VGHH40 pKa = 5.97DD41 pKa = 3.24HH42 pKa = 6.69LQGFIQFKK50 pKa = 10.55SRR52 pKa = 11.84NTTMGQCKK60 pKa = 9.83AQFNGLSPHH69 pKa = 6.95LEE71 pKa = 3.61IARR74 pKa = 11.84DD75 pKa = 3.61VNKK78 pKa = 10.54AQEE81 pKa = 4.13YY82 pKa = 8.8TMKK85 pKa = 10.39EE86 pKa = 4.06DD87 pKa = 3.49TRR89 pKa = 11.84IAGPWEE95 pKa = 3.9YY96 pKa = 11.4GVMIKK101 pKa = 10.33KK102 pKa = 10.28GSHH105 pKa = 4.91KK106 pKa = 10.1RR107 pKa = 11.84KK108 pKa = 9.94LMEE111 pKa = 4.6KK112 pKa = 10.26FEE114 pKa = 4.53EE115 pKa = 4.82DD116 pKa = 3.56PEE118 pKa = 4.16EE119 pKa = 4.08MKK121 pKa = 10.7IADD124 pKa = 3.53PSLYY128 pKa = 10.0RR129 pKa = 11.84RR130 pKa = 11.84CLTRR134 pKa = 11.84KK135 pKa = 8.7MSEE138 pKa = 4.1EE139 pKa = 3.94QRR141 pKa = 11.84SSAEE145 pKa = 3.47WNYY148 pKa = 11.24DD149 pKa = 3.23LRR151 pKa = 11.84PWQEE155 pKa = 3.37AVIQEE160 pKa = 4.5LEE162 pKa = 4.23LPPDD166 pKa = 3.34YY167 pKa = 10.99RR168 pKa = 11.84KK169 pKa = 10.23IIWVYY174 pKa = 10.71GPGGNEE180 pKa = 4.03GKK182 pKa = 8.49STFARR187 pKa = 11.84HH188 pKa = 6.0LSLKK192 pKa = 9.77EE193 pKa = 3.4GWGYY197 pKa = 10.96VPGGRR202 pKa = 11.84TQDD205 pKa = 3.09MMHH208 pKa = 7.46IITADD213 pKa = 3.75PKK215 pKa = 8.93NHH217 pKa = 5.83WVFDD221 pKa = 3.86VPRR224 pKa = 11.84VSSEE228 pKa = 3.72YY229 pKa = 10.22VNYY232 pKa = 10.52GVIEE236 pKa = 4.05QVKK239 pKa = 9.26NRR241 pKa = 11.84VMVNTKK247 pKa = 10.11YY248 pKa = 10.08EE249 pKa = 4.13PCVMRR254 pKa = 11.84DD255 pKa = 3.5DD256 pKa = 3.89NHH258 pKa = 6.28PVHH261 pKa = 6.57VVVFANCLPDD271 pKa = 3.58VTKK274 pKa = 10.94LSEE277 pKa = 4.03DD278 pKa = 3.48RR279 pKa = 11.84IKK281 pKa = 10.49IIKK284 pKa = 9.41CC285 pKa = 3.38
MM1 pKa = 7.98PSQKK5 pKa = 8.99STCWVFTLNFEE16 pKa = 4.51GDD18 pKa = 3.83PPTLTFDD25 pKa = 5.33DD26 pKa = 4.08RR27 pKa = 11.84VQYY30 pKa = 11.05ASWQHH35 pKa = 5.1EE36 pKa = 4.64RR37 pKa = 11.84VGHH40 pKa = 5.97DD41 pKa = 3.24HH42 pKa = 6.69LQGFIQFKK50 pKa = 10.55SRR52 pKa = 11.84NTTMGQCKK60 pKa = 9.83AQFNGLSPHH69 pKa = 6.95LEE71 pKa = 3.61IARR74 pKa = 11.84DD75 pKa = 3.61VNKK78 pKa = 10.54AQEE81 pKa = 4.13YY82 pKa = 8.8TMKK85 pKa = 10.39EE86 pKa = 4.06DD87 pKa = 3.49TRR89 pKa = 11.84IAGPWEE95 pKa = 3.9YY96 pKa = 11.4GVMIKK101 pKa = 10.33KK102 pKa = 10.28GSHH105 pKa = 4.91KK106 pKa = 10.1RR107 pKa = 11.84KK108 pKa = 9.94LMEE111 pKa = 4.6KK112 pKa = 10.26FEE114 pKa = 4.53EE115 pKa = 4.82DD116 pKa = 3.56PEE118 pKa = 4.16EE119 pKa = 4.08MKK121 pKa = 10.7IADD124 pKa = 3.53PSLYY128 pKa = 10.0RR129 pKa = 11.84RR130 pKa = 11.84CLTRR134 pKa = 11.84KK135 pKa = 8.7MSEE138 pKa = 4.1EE139 pKa = 3.94QRR141 pKa = 11.84SSAEE145 pKa = 3.47WNYY148 pKa = 11.24DD149 pKa = 3.23LRR151 pKa = 11.84PWQEE155 pKa = 3.37AVIQEE160 pKa = 4.5LEE162 pKa = 4.23LPPDD166 pKa = 3.34YY167 pKa = 10.99RR168 pKa = 11.84KK169 pKa = 10.23IIWVYY174 pKa = 10.71GPGGNEE180 pKa = 4.03GKK182 pKa = 8.49STFARR187 pKa = 11.84HH188 pKa = 6.0LSLKK192 pKa = 9.77EE193 pKa = 3.4GWGYY197 pKa = 10.96VPGGRR202 pKa = 11.84TQDD205 pKa = 3.09MMHH208 pKa = 7.46IITADD213 pKa = 3.75PKK215 pKa = 8.93NHH217 pKa = 5.83WVFDD221 pKa = 3.86VPRR224 pKa = 11.84VSSEE228 pKa = 3.72YY229 pKa = 10.22VNYY232 pKa = 10.52GVIEE236 pKa = 4.05QVKK239 pKa = 9.26NRR241 pKa = 11.84VMVNTKK247 pKa = 10.11YY248 pKa = 10.08EE249 pKa = 4.13PCVMRR254 pKa = 11.84DD255 pKa = 3.5DD256 pKa = 3.89NHH258 pKa = 6.28PVHH261 pKa = 6.57VVVFANCLPDD271 pKa = 3.58VTKK274 pKa = 10.94LSEE277 pKa = 4.03DD278 pKa = 3.48RR279 pKa = 11.84IKK281 pKa = 10.49IIKK284 pKa = 9.41CC285 pKa = 3.38
Molecular weight: 33.26 kDa
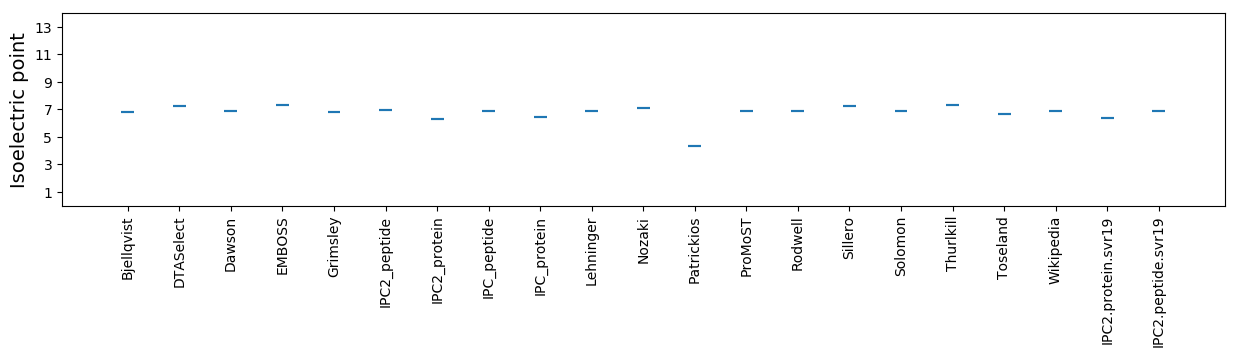
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5YSH3|A0A1W5YSH3_9VIRU Satellite replication initiator protein OS=Sophora alopecuroides yellow stunt alphasatellite 8 OX=1980173 PE=4 SV=1
MM1 pKa = 7.98PSQKK5 pKa = 8.99STCWVFTLNFEE16 pKa = 4.51GDD18 pKa = 3.83PPTLTFDD25 pKa = 5.33DD26 pKa = 4.08RR27 pKa = 11.84VQYY30 pKa = 11.05ASWQHH35 pKa = 5.1EE36 pKa = 4.64RR37 pKa = 11.84VGHH40 pKa = 5.97DD41 pKa = 3.24HH42 pKa = 6.69LQGFIQFKK50 pKa = 10.55SRR52 pKa = 11.84NTTMGQCKK60 pKa = 9.83AQFNGLSPHH69 pKa = 6.95LEE71 pKa = 3.61IARR74 pKa = 11.84DD75 pKa = 3.61VNKK78 pKa = 10.54AQEE81 pKa = 4.13YY82 pKa = 8.8TMKK85 pKa = 10.39EE86 pKa = 4.06DD87 pKa = 3.49TRR89 pKa = 11.84IAGPWEE95 pKa = 3.9YY96 pKa = 11.4GVMIKK101 pKa = 10.33KK102 pKa = 10.28GSHH105 pKa = 4.91KK106 pKa = 10.1RR107 pKa = 11.84KK108 pKa = 9.94LMEE111 pKa = 4.6KK112 pKa = 10.26FEE114 pKa = 4.53EE115 pKa = 4.82DD116 pKa = 3.56PEE118 pKa = 4.16EE119 pKa = 4.08MKK121 pKa = 10.7IADD124 pKa = 3.53PSLYY128 pKa = 10.0RR129 pKa = 11.84RR130 pKa = 11.84CLTRR134 pKa = 11.84KK135 pKa = 8.7MSEE138 pKa = 4.1EE139 pKa = 3.94QRR141 pKa = 11.84SSAEE145 pKa = 3.47WNYY148 pKa = 11.24DD149 pKa = 3.23LRR151 pKa = 11.84PWQEE155 pKa = 3.37AVIQEE160 pKa = 4.5LEE162 pKa = 4.23LPPDD166 pKa = 3.34YY167 pKa = 10.99RR168 pKa = 11.84KK169 pKa = 10.23IIWVYY174 pKa = 10.71GPGGNEE180 pKa = 4.03GKK182 pKa = 8.49STFARR187 pKa = 11.84HH188 pKa = 6.0LSLKK192 pKa = 9.77EE193 pKa = 3.4GWGYY197 pKa = 10.96VPGGRR202 pKa = 11.84TQDD205 pKa = 3.09MMHH208 pKa = 7.46IITADD213 pKa = 3.75PKK215 pKa = 8.93NHH217 pKa = 5.83WVFDD221 pKa = 3.86VPRR224 pKa = 11.84VSSEE228 pKa = 3.72YY229 pKa = 10.22VNYY232 pKa = 10.52GVIEE236 pKa = 4.05QVKK239 pKa = 9.26NRR241 pKa = 11.84VMVNTKK247 pKa = 10.11YY248 pKa = 10.08EE249 pKa = 4.13PCVMRR254 pKa = 11.84DD255 pKa = 3.5DD256 pKa = 3.89NHH258 pKa = 6.28PVHH261 pKa = 6.57VVVFANCLPDD271 pKa = 3.58VTKK274 pKa = 10.94LSEE277 pKa = 4.03DD278 pKa = 3.48RR279 pKa = 11.84IKK281 pKa = 10.49IIKK284 pKa = 9.41CC285 pKa = 3.38
MM1 pKa = 7.98PSQKK5 pKa = 8.99STCWVFTLNFEE16 pKa = 4.51GDD18 pKa = 3.83PPTLTFDD25 pKa = 5.33DD26 pKa = 4.08RR27 pKa = 11.84VQYY30 pKa = 11.05ASWQHH35 pKa = 5.1EE36 pKa = 4.64RR37 pKa = 11.84VGHH40 pKa = 5.97DD41 pKa = 3.24HH42 pKa = 6.69LQGFIQFKK50 pKa = 10.55SRR52 pKa = 11.84NTTMGQCKK60 pKa = 9.83AQFNGLSPHH69 pKa = 6.95LEE71 pKa = 3.61IARR74 pKa = 11.84DD75 pKa = 3.61VNKK78 pKa = 10.54AQEE81 pKa = 4.13YY82 pKa = 8.8TMKK85 pKa = 10.39EE86 pKa = 4.06DD87 pKa = 3.49TRR89 pKa = 11.84IAGPWEE95 pKa = 3.9YY96 pKa = 11.4GVMIKK101 pKa = 10.33KK102 pKa = 10.28GSHH105 pKa = 4.91KK106 pKa = 10.1RR107 pKa = 11.84KK108 pKa = 9.94LMEE111 pKa = 4.6KK112 pKa = 10.26FEE114 pKa = 4.53EE115 pKa = 4.82DD116 pKa = 3.56PEE118 pKa = 4.16EE119 pKa = 4.08MKK121 pKa = 10.7IADD124 pKa = 3.53PSLYY128 pKa = 10.0RR129 pKa = 11.84RR130 pKa = 11.84CLTRR134 pKa = 11.84KK135 pKa = 8.7MSEE138 pKa = 4.1EE139 pKa = 3.94QRR141 pKa = 11.84SSAEE145 pKa = 3.47WNYY148 pKa = 11.24DD149 pKa = 3.23LRR151 pKa = 11.84PWQEE155 pKa = 3.37AVIQEE160 pKa = 4.5LEE162 pKa = 4.23LPPDD166 pKa = 3.34YY167 pKa = 10.99RR168 pKa = 11.84KK169 pKa = 10.23IIWVYY174 pKa = 10.71GPGGNEE180 pKa = 4.03GKK182 pKa = 8.49STFARR187 pKa = 11.84HH188 pKa = 6.0LSLKK192 pKa = 9.77EE193 pKa = 3.4GWGYY197 pKa = 10.96VPGGRR202 pKa = 11.84TQDD205 pKa = 3.09MMHH208 pKa = 7.46IITADD213 pKa = 3.75PKK215 pKa = 8.93NHH217 pKa = 5.83WVFDD221 pKa = 3.86VPRR224 pKa = 11.84VSSEE228 pKa = 3.72YY229 pKa = 10.22VNYY232 pKa = 10.52GVIEE236 pKa = 4.05QVKK239 pKa = 9.26NRR241 pKa = 11.84VMVNTKK247 pKa = 10.11YY248 pKa = 10.08EE249 pKa = 4.13PCVMRR254 pKa = 11.84DD255 pKa = 3.5DD256 pKa = 3.89NHH258 pKa = 6.28PVHH261 pKa = 6.57VVVFANCLPDD271 pKa = 3.58VTKK274 pKa = 10.94LSEE277 pKa = 4.03DD278 pKa = 3.48RR279 pKa = 11.84IKK281 pKa = 10.49IIKK284 pKa = 9.41CC285 pKa = 3.38
Molecular weight: 33.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
285 |
285 |
285 |
285.0 |
33.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.86 ± 0.0 | 2.105 ± 0.0 |
5.965 ± 0.0 | 8.07 ± 0.0 |
3.509 ± 0.0 | 5.965 ± 0.0 |
3.509 ± 0.0 | 4.912 ± 0.0 |
7.368 ± 0.0 | 5.263 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.86 ± 0.0 | 4.211 ± 0.0 |
5.965 ± 0.0 | 4.561 ± 0.0 |
6.316 ± 0.0 | 5.263 ± 0.0 |
4.912 ± 0.0 | 7.719 ± 0.0 |
2.807 ± 0.0 | 3.86 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
