
Corynespora cassiicola Philippines
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Pleosporomycetidae; Pleosporales; Corynesporascaceae; Corynespora; Corynespora cassiicola
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
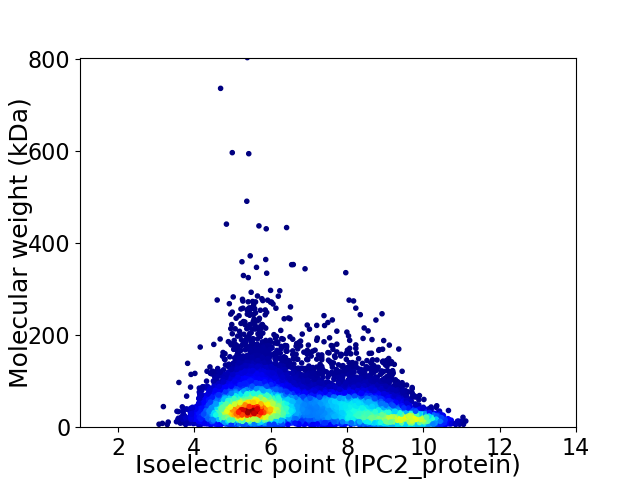
Virtual 2D-PAGE plot for 17125 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
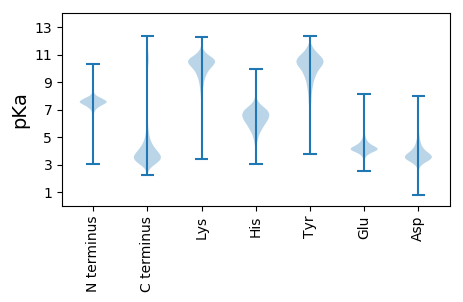
Protein with the lowest isoelectric point:
>tr|A0A2T2N800|A0A2T2N800_CORCC Short chain dehydrogenase OS=Corynespora cassiicola Philippines OX=1448308 GN=BS50DRAFT_651787 PE=4 SV=1
MM1 pKa = 7.57GGFSRR6 pKa = 11.84ISALLLAAGYY16 pKa = 10.04LGSEE20 pKa = 4.44VAAAAVPAVPVPSAPQTDD38 pKa = 3.68RR39 pKa = 11.84VLRR42 pKa = 11.84AATRR46 pKa = 11.84RR47 pKa = 11.84SNLHH51 pKa = 5.55RR52 pKa = 11.84RR53 pKa = 11.84GLRR56 pKa = 11.84IEE58 pKa = 4.24KK59 pKa = 10.52KK60 pKa = 10.19FGSEE64 pKa = 3.23LAYY67 pKa = 9.86IEE69 pKa = 4.68EE70 pKa = 4.49EE71 pKa = 4.1NGLSGRR77 pKa = 11.84PVFASHH83 pKa = 5.28VTVGSKK89 pKa = 10.54KK90 pKa = 10.24PILNLEE96 pKa = 4.3EE97 pKa = 5.25IEE99 pKa = 6.45DD100 pKa = 3.8YY101 pKa = 11.21LQDD104 pKa = 3.79VKK106 pKa = 11.13CGSDD110 pKa = 3.04GKK112 pKa = 10.3MKK114 pKa = 10.74LVFKK118 pKa = 10.78DD119 pKa = 4.43RR120 pKa = 11.84IAARR124 pKa = 11.84DD125 pKa = 3.56ARR127 pKa = 11.84LACSDD132 pKa = 3.92EE133 pKa = 4.64DD134 pKa = 3.62GGLIITSHH142 pKa = 6.47EE143 pKa = 4.13GCNVDD148 pKa = 3.97GEE150 pKa = 4.34RR151 pKa = 11.84SVYY154 pKa = 9.44SVKK157 pKa = 10.74DD158 pKa = 2.96IAFADD163 pKa = 3.97DD164 pKa = 3.91EE165 pKa = 4.63NALEE169 pKa = 4.03LAVAEE174 pKa = 4.8KK175 pKa = 8.96MWKK178 pKa = 10.12DD179 pKa = 3.36AFDD182 pKa = 4.02HH183 pKa = 6.5FDD185 pKa = 2.87ISFGHH190 pKa = 5.54TTDD193 pKa = 2.96KK194 pKa = 10.7FHH196 pKa = 6.83YY197 pKa = 7.51RR198 pKa = 11.84THH200 pKa = 7.13RR201 pKa = 11.84DD202 pKa = 3.16FARR205 pKa = 11.84RR206 pKa = 11.84KK207 pKa = 8.07RR208 pKa = 11.84QEE210 pKa = 3.82AGEE213 pKa = 4.12PDD215 pKa = 3.59ITPQPVEE222 pKa = 4.47IPTDD226 pKa = 3.31IPDD229 pKa = 4.08DD230 pKa = 4.23VNSVEE235 pKa = 6.32FDD237 pKa = 3.79LTWEE241 pKa = 4.7KK242 pKa = 10.21IDD244 pKa = 3.76EE245 pKa = 4.37VFSVSDD251 pKa = 3.6FTAGLEE257 pKa = 4.09QFVNVPTPADD267 pKa = 3.57LPIEE271 pKa = 4.44LGCTEE276 pKa = 5.49CSTQGTITLSQGAFEE291 pKa = 4.79IDD293 pKa = 3.41VGEE296 pKa = 4.56LDD298 pKa = 3.81VVPDD302 pKa = 4.35FLQGGDD308 pKa = 3.77DD309 pKa = 4.02GKK311 pKa = 10.72DD312 pKa = 2.86ISSVITGGFIEE323 pKa = 5.06LKK325 pKa = 9.01ANSVFGHH332 pKa = 6.6FDD334 pKa = 3.41LFARR338 pKa = 11.84PRR340 pKa = 11.84AAGSFEE346 pKa = 3.64ITLFSLPVLGFTIPNIGRR364 pKa = 11.84AGAVFEE370 pKa = 5.2AGLSADD376 pKa = 3.64YY377 pKa = 10.78DD378 pKa = 4.05VEE380 pKa = 6.01GGLDD384 pKa = 3.65LSYY387 pKa = 11.16GIDD390 pKa = 3.36VTVPDD395 pKa = 4.68DD396 pKa = 3.47ASMRR400 pKa = 11.84IEE402 pKa = 4.98LGDD405 pKa = 3.71LKK407 pKa = 11.01KK408 pKa = 10.99SGVSGFAEE416 pKa = 4.39STLEE420 pKa = 3.91PLPFSANVTDD430 pKa = 4.14VDD432 pKa = 4.0ILLGLAFVPTIPIGFMFADD451 pKa = 3.95KK452 pKa = 10.82LDD454 pKa = 3.99ASVTVSMDD462 pKa = 3.89LPRR465 pKa = 11.84LDD467 pKa = 4.87ALITAKK473 pKa = 10.71PNGDD477 pKa = 3.76CKK479 pKa = 11.04SILGGNSTSVASLGNSTNGDD499 pKa = 4.13LSPVIIVDD507 pKa = 3.66ANVSIVLDD515 pKa = 3.65VGADD519 pKa = 3.59FSFPILPIDD528 pKa = 4.68PIGTAAEE535 pKa = 3.92IFSTKK540 pKa = 10.19FEE542 pKa = 5.01LGTRR546 pKa = 11.84CLAPGEE552 pKa = 4.48GFAQATGAAAAGVGNQLPGFVLTAGYY578 pKa = 10.28GDD580 pKa = 3.84SASTCGGSTVTVYY593 pKa = 10.43QPPPTGGAQYY603 pKa = 10.53PPYY606 pKa = 9.71EE607 pKa = 4.24PSAPVIEE614 pKa = 4.98PPVITPPPCSGYY626 pKa = 10.64DD627 pKa = 3.54CPDD630 pKa = 3.51SPEE633 pKa = 4.66SPPSPPVQSEE643 pKa = 4.35PNCNGYY649 pKa = 9.61EE650 pKa = 4.27CPKK653 pKa = 10.13PPSPPVEE660 pKa = 4.59SNPPPPPAEE669 pKa = 4.36SDD671 pKa = 3.64PGCNGYY677 pKa = 10.11DD678 pKa = 3.78CPPPVAPPQSEE689 pKa = 4.42PPSPPQSEE697 pKa = 4.28PVSPPVEE704 pKa = 4.24SDD706 pKa = 3.38PGCNGYY712 pKa = 9.88DD713 pKa = 3.94CPQQPPVEE721 pKa = 4.4PPIEE725 pKa = 4.39PPVTTTPCDD734 pKa = 3.58SSTLPPATEE743 pKa = 4.09PPKK746 pKa = 10.33TDD748 pKa = 3.33SPPPEE753 pKa = 4.68IPSAPVEE760 pKa = 4.44PPVSTAPPSPPEE772 pKa = 4.15QPSSPPEE779 pKa = 4.1SPPEE783 pKa = 4.1QPSSPPEE790 pKa = 4.09NPPEE794 pKa = 4.25QPSSPPEE801 pKa = 4.12NPPVEE806 pKa = 4.4PPKK809 pKa = 8.76TTAPPPPPSTDD820 pKa = 3.16VPEE823 pKa = 4.8PPSTTVCDD831 pKa = 4.57DD832 pKa = 3.69YY833 pKa = 11.9SCNEE837 pKa = 4.05SPEE840 pKa = 4.28NPPSQPAIPPMEE852 pKa = 4.67PPSTTEE858 pKa = 3.66SSEE861 pKa = 4.1QPTPPPEE868 pKa = 4.39NPPEE872 pKa = 4.27VPSVPVEE879 pKa = 4.15PPVSSSMVEE888 pKa = 4.16PPPEE892 pKa = 4.66APTPPPCDD900 pKa = 4.15GYY902 pKa = 11.4DD903 pKa = 3.46CTEE906 pKa = 4.17TPDD909 pKa = 3.83VPSQPVEE916 pKa = 4.28PPMSTPPPPPVDD928 pKa = 3.79EE929 pKa = 4.78PSVPAEE935 pKa = 4.31PPVSPPSTGDD945 pKa = 3.15IFPTPPPCDD954 pKa = 4.39GYY956 pKa = 11.32DD957 pKa = 3.44CPEE960 pKa = 4.3NPDD963 pKa = 3.76VPSQPAEE970 pKa = 4.25PPVSTPTPPPTVEE983 pKa = 4.3EE984 pKa = 4.16PSEE987 pKa = 4.05PVNPPVSPPSTGDD1000 pKa = 3.27IVPTPPPEE1008 pKa = 4.37NPEE1011 pKa = 4.28SPPEE1015 pKa = 3.98QPPTTSAPEE1024 pKa = 3.86CDD1026 pKa = 4.52GYY1028 pKa = 11.58GCEE1031 pKa = 5.1PPTEE1035 pKa = 4.25TPPPSGDD1042 pKa = 3.56EE1043 pKa = 3.73PSKK1046 pKa = 10.61PIEE1049 pKa = 4.65PPVSTPPTEE1058 pKa = 4.79PPTEE1062 pKa = 4.21TPPPCDD1068 pKa = 3.34GDD1070 pKa = 4.38CPQEE1074 pKa = 4.45PPAEE1078 pKa = 4.51PPVSTPVTEE1087 pKa = 5.1PPTEE1091 pKa = 4.17TPPPCDD1097 pKa = 3.46GDD1099 pKa = 4.49CPTEE1103 pKa = 4.52PPTEE1107 pKa = 4.21TPPPCDD1113 pKa = 5.01GYY1115 pKa = 11.39DD1116 pKa = 3.9CNNSQPPTEE1125 pKa = 5.15PPTEE1129 pKa = 4.09TPPPGGYY1136 pKa = 9.76EE1137 pKa = 3.73PSKK1140 pKa = 9.94PAEE1143 pKa = 4.6PPVSTPPTEE1152 pKa = 4.79PPTEE1156 pKa = 4.21TPPPCDD1162 pKa = 3.46GDD1164 pKa = 4.49CPTEE1168 pKa = 4.52PPTEE1172 pKa = 4.21TPPPCDD1178 pKa = 4.67GYY1180 pKa = 11.38DD1181 pKa = 3.61CPTEE1185 pKa = 4.07SSPPCSGPDD1194 pKa = 3.57CPSEE1198 pKa = 4.37PPVQTPPPSAEE1209 pKa = 4.37TPCTTSSTQTITLSTGGTGGGGYY1232 pKa = 9.81PNTTAGGYY1240 pKa = 9.5GHH1242 pKa = 7.26TPPSYY1247 pKa = 10.29PPATTMVEE1255 pKa = 4.12VPPPYY1260 pKa = 10.21GGYY1263 pKa = 9.43PPPASHH1269 pKa = 6.88TGNMTMQTGFLDD1281 pKa = 4.59GPTGTRR1287 pKa = 11.84PPMQTTSPAQFTGAAVPRR1305 pKa = 11.84AVPIAVDD1312 pKa = 3.54AKK1314 pKa = 10.16GLGWQGAVLVMSLFAGGFMLLL1335 pKa = 3.88
MM1 pKa = 7.57GGFSRR6 pKa = 11.84ISALLLAAGYY16 pKa = 10.04LGSEE20 pKa = 4.44VAAAAVPAVPVPSAPQTDD38 pKa = 3.68RR39 pKa = 11.84VLRR42 pKa = 11.84AATRR46 pKa = 11.84RR47 pKa = 11.84SNLHH51 pKa = 5.55RR52 pKa = 11.84RR53 pKa = 11.84GLRR56 pKa = 11.84IEE58 pKa = 4.24KK59 pKa = 10.52KK60 pKa = 10.19FGSEE64 pKa = 3.23LAYY67 pKa = 9.86IEE69 pKa = 4.68EE70 pKa = 4.49EE71 pKa = 4.1NGLSGRR77 pKa = 11.84PVFASHH83 pKa = 5.28VTVGSKK89 pKa = 10.54KK90 pKa = 10.24PILNLEE96 pKa = 4.3EE97 pKa = 5.25IEE99 pKa = 6.45DD100 pKa = 3.8YY101 pKa = 11.21LQDD104 pKa = 3.79VKK106 pKa = 11.13CGSDD110 pKa = 3.04GKK112 pKa = 10.3MKK114 pKa = 10.74LVFKK118 pKa = 10.78DD119 pKa = 4.43RR120 pKa = 11.84IAARR124 pKa = 11.84DD125 pKa = 3.56ARR127 pKa = 11.84LACSDD132 pKa = 3.92EE133 pKa = 4.64DD134 pKa = 3.62GGLIITSHH142 pKa = 6.47EE143 pKa = 4.13GCNVDD148 pKa = 3.97GEE150 pKa = 4.34RR151 pKa = 11.84SVYY154 pKa = 9.44SVKK157 pKa = 10.74DD158 pKa = 2.96IAFADD163 pKa = 3.97DD164 pKa = 3.91EE165 pKa = 4.63NALEE169 pKa = 4.03LAVAEE174 pKa = 4.8KK175 pKa = 8.96MWKK178 pKa = 10.12DD179 pKa = 3.36AFDD182 pKa = 4.02HH183 pKa = 6.5FDD185 pKa = 2.87ISFGHH190 pKa = 5.54TTDD193 pKa = 2.96KK194 pKa = 10.7FHH196 pKa = 6.83YY197 pKa = 7.51RR198 pKa = 11.84THH200 pKa = 7.13RR201 pKa = 11.84DD202 pKa = 3.16FARR205 pKa = 11.84RR206 pKa = 11.84KK207 pKa = 8.07RR208 pKa = 11.84QEE210 pKa = 3.82AGEE213 pKa = 4.12PDD215 pKa = 3.59ITPQPVEE222 pKa = 4.47IPTDD226 pKa = 3.31IPDD229 pKa = 4.08DD230 pKa = 4.23VNSVEE235 pKa = 6.32FDD237 pKa = 3.79LTWEE241 pKa = 4.7KK242 pKa = 10.21IDD244 pKa = 3.76EE245 pKa = 4.37VFSVSDD251 pKa = 3.6FTAGLEE257 pKa = 4.09QFVNVPTPADD267 pKa = 3.57LPIEE271 pKa = 4.44LGCTEE276 pKa = 5.49CSTQGTITLSQGAFEE291 pKa = 4.79IDD293 pKa = 3.41VGEE296 pKa = 4.56LDD298 pKa = 3.81VVPDD302 pKa = 4.35FLQGGDD308 pKa = 3.77DD309 pKa = 4.02GKK311 pKa = 10.72DD312 pKa = 2.86ISSVITGGFIEE323 pKa = 5.06LKK325 pKa = 9.01ANSVFGHH332 pKa = 6.6FDD334 pKa = 3.41LFARR338 pKa = 11.84PRR340 pKa = 11.84AAGSFEE346 pKa = 3.64ITLFSLPVLGFTIPNIGRR364 pKa = 11.84AGAVFEE370 pKa = 5.2AGLSADD376 pKa = 3.64YY377 pKa = 10.78DD378 pKa = 4.05VEE380 pKa = 6.01GGLDD384 pKa = 3.65LSYY387 pKa = 11.16GIDD390 pKa = 3.36VTVPDD395 pKa = 4.68DD396 pKa = 3.47ASMRR400 pKa = 11.84IEE402 pKa = 4.98LGDD405 pKa = 3.71LKK407 pKa = 11.01KK408 pKa = 10.99SGVSGFAEE416 pKa = 4.39STLEE420 pKa = 3.91PLPFSANVTDD430 pKa = 4.14VDD432 pKa = 4.0ILLGLAFVPTIPIGFMFADD451 pKa = 3.95KK452 pKa = 10.82LDD454 pKa = 3.99ASVTVSMDD462 pKa = 3.89LPRR465 pKa = 11.84LDD467 pKa = 4.87ALITAKK473 pKa = 10.71PNGDD477 pKa = 3.76CKK479 pKa = 11.04SILGGNSTSVASLGNSTNGDD499 pKa = 4.13LSPVIIVDD507 pKa = 3.66ANVSIVLDD515 pKa = 3.65VGADD519 pKa = 3.59FSFPILPIDD528 pKa = 4.68PIGTAAEE535 pKa = 3.92IFSTKK540 pKa = 10.19FEE542 pKa = 5.01LGTRR546 pKa = 11.84CLAPGEE552 pKa = 4.48GFAQATGAAAAGVGNQLPGFVLTAGYY578 pKa = 10.28GDD580 pKa = 3.84SASTCGGSTVTVYY593 pKa = 10.43QPPPTGGAQYY603 pKa = 10.53PPYY606 pKa = 9.71EE607 pKa = 4.24PSAPVIEE614 pKa = 4.98PPVITPPPCSGYY626 pKa = 10.64DD627 pKa = 3.54CPDD630 pKa = 3.51SPEE633 pKa = 4.66SPPSPPVQSEE643 pKa = 4.35PNCNGYY649 pKa = 9.61EE650 pKa = 4.27CPKK653 pKa = 10.13PPSPPVEE660 pKa = 4.59SNPPPPPAEE669 pKa = 4.36SDD671 pKa = 3.64PGCNGYY677 pKa = 10.11DD678 pKa = 3.78CPPPVAPPQSEE689 pKa = 4.42PPSPPQSEE697 pKa = 4.28PVSPPVEE704 pKa = 4.24SDD706 pKa = 3.38PGCNGYY712 pKa = 9.88DD713 pKa = 3.94CPQQPPVEE721 pKa = 4.4PPIEE725 pKa = 4.39PPVTTTPCDD734 pKa = 3.58SSTLPPATEE743 pKa = 4.09PPKK746 pKa = 10.33TDD748 pKa = 3.33SPPPEE753 pKa = 4.68IPSAPVEE760 pKa = 4.44PPVSTAPPSPPEE772 pKa = 4.15QPSSPPEE779 pKa = 4.1SPPEE783 pKa = 4.1QPSSPPEE790 pKa = 4.09NPPEE794 pKa = 4.25QPSSPPEE801 pKa = 4.12NPPVEE806 pKa = 4.4PPKK809 pKa = 8.76TTAPPPPPSTDD820 pKa = 3.16VPEE823 pKa = 4.8PPSTTVCDD831 pKa = 4.57DD832 pKa = 3.69YY833 pKa = 11.9SCNEE837 pKa = 4.05SPEE840 pKa = 4.28NPPSQPAIPPMEE852 pKa = 4.67PPSTTEE858 pKa = 3.66SSEE861 pKa = 4.1QPTPPPEE868 pKa = 4.39NPPEE872 pKa = 4.27VPSVPVEE879 pKa = 4.15PPVSSSMVEE888 pKa = 4.16PPPEE892 pKa = 4.66APTPPPCDD900 pKa = 4.15GYY902 pKa = 11.4DD903 pKa = 3.46CTEE906 pKa = 4.17TPDD909 pKa = 3.83VPSQPVEE916 pKa = 4.28PPMSTPPPPPVDD928 pKa = 3.79EE929 pKa = 4.78PSVPAEE935 pKa = 4.31PPVSPPSTGDD945 pKa = 3.15IFPTPPPCDD954 pKa = 4.39GYY956 pKa = 11.32DD957 pKa = 3.44CPEE960 pKa = 4.3NPDD963 pKa = 3.76VPSQPAEE970 pKa = 4.25PPVSTPTPPPTVEE983 pKa = 4.3EE984 pKa = 4.16PSEE987 pKa = 4.05PVNPPVSPPSTGDD1000 pKa = 3.27IVPTPPPEE1008 pKa = 4.37NPEE1011 pKa = 4.28SPPEE1015 pKa = 3.98QPPTTSAPEE1024 pKa = 3.86CDD1026 pKa = 4.52GYY1028 pKa = 11.58GCEE1031 pKa = 5.1PPTEE1035 pKa = 4.25TPPPSGDD1042 pKa = 3.56EE1043 pKa = 3.73PSKK1046 pKa = 10.61PIEE1049 pKa = 4.65PPVSTPPTEE1058 pKa = 4.79PPTEE1062 pKa = 4.21TPPPCDD1068 pKa = 3.34GDD1070 pKa = 4.38CPQEE1074 pKa = 4.45PPAEE1078 pKa = 4.51PPVSTPVTEE1087 pKa = 5.1PPTEE1091 pKa = 4.17TPPPCDD1097 pKa = 3.46GDD1099 pKa = 4.49CPTEE1103 pKa = 4.52PPTEE1107 pKa = 4.21TPPPCDD1113 pKa = 5.01GYY1115 pKa = 11.39DD1116 pKa = 3.9CNNSQPPTEE1125 pKa = 5.15PPTEE1129 pKa = 4.09TPPPGGYY1136 pKa = 9.76EE1137 pKa = 3.73PSKK1140 pKa = 9.94PAEE1143 pKa = 4.6PPVSTPPTEE1152 pKa = 4.79PPTEE1156 pKa = 4.21TPPPCDD1162 pKa = 3.46GDD1164 pKa = 4.49CPTEE1168 pKa = 4.52PPTEE1172 pKa = 4.21TPPPCDD1178 pKa = 4.67GYY1180 pKa = 11.38DD1181 pKa = 3.61CPTEE1185 pKa = 4.07SSPPCSGPDD1194 pKa = 3.57CPSEE1198 pKa = 4.37PPVQTPPPSAEE1209 pKa = 4.37TPCTTSSTQTITLSTGGTGGGGYY1232 pKa = 9.81PNTTAGGYY1240 pKa = 9.5GHH1242 pKa = 7.26TPPSYY1247 pKa = 10.29PPATTMVEE1255 pKa = 4.12VPPPYY1260 pKa = 10.21GGYY1263 pKa = 9.43PPPASHH1269 pKa = 6.88TGNMTMQTGFLDD1281 pKa = 4.59GPTGTRR1287 pKa = 11.84PPMQTTSPAQFTGAAVPRR1305 pKa = 11.84AVPIAVDD1312 pKa = 3.54AKK1314 pKa = 10.16GLGWQGAVLVMSLFAGGFMLLL1335 pKa = 3.88
Molecular weight: 138.64 kDa
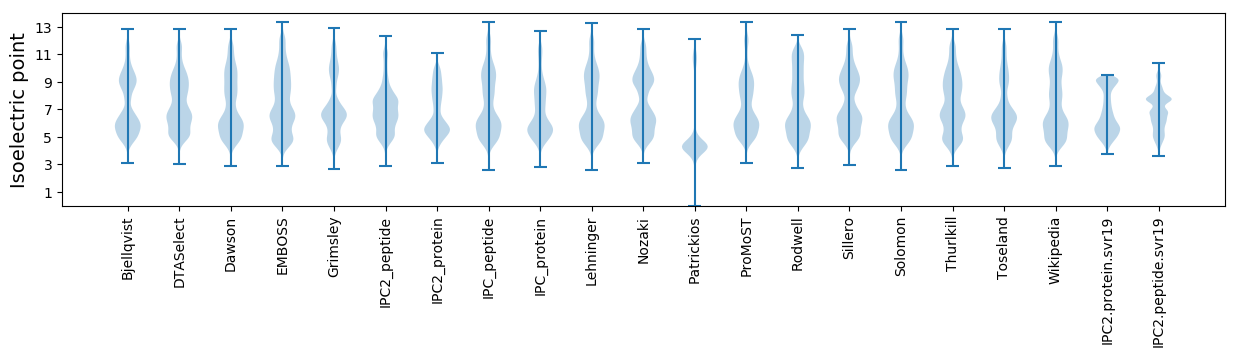
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T2NI65|A0A2T2NI65_CORCC Uncharacterized protein OS=Corynespora cassiicola Philippines OX=1448308 GN=BS50DRAFT_51737 PE=4 SV=1
MM1 pKa = 7.34AVAVAAGGGLTAALRR16 pKa = 11.84PPSKK20 pKa = 10.37PSPTTTASRR29 pKa = 11.84ASQALRR35 pKa = 11.84LPSARR40 pKa = 11.84CPLPSALSQPSRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84ASGASPATAPPLPTASIKK72 pKa = 9.8PANGPSALAPSRR84 pKa = 11.84PRR86 pKa = 11.84RR87 pKa = 11.84AARR90 pKa = 11.84LAMAARR96 pKa = 11.84APPPTAARR104 pKa = 11.84HH105 pKa = 5.13PPSAIRR111 pKa = 11.84HH112 pKa = 5.38PPSAIHH118 pKa = 6.74RR119 pKa = 11.84PSQTRR124 pKa = 11.84LPSQSRR130 pKa = 11.84LGG132 pKa = 3.55
MM1 pKa = 7.34AVAVAAGGGLTAALRR16 pKa = 11.84PPSKK20 pKa = 10.37PSPTTTASRR29 pKa = 11.84ASQALRR35 pKa = 11.84LPSARR40 pKa = 11.84CPLPSALSQPSRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84ASGASPATAPPLPTASIKK72 pKa = 9.8PANGPSALAPSRR84 pKa = 11.84PRR86 pKa = 11.84RR87 pKa = 11.84AARR90 pKa = 11.84LAMAARR96 pKa = 11.84APPPTAARR104 pKa = 11.84HH105 pKa = 5.13PPSAIRR111 pKa = 11.84HH112 pKa = 5.38PPSAIHH118 pKa = 6.74RR119 pKa = 11.84PSQTRR124 pKa = 11.84LPSQSRR130 pKa = 11.84LGG132 pKa = 3.55
Molecular weight: 13.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7195570 |
49 |
7227 |
420.2 |
46.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.65 ± 0.017 | 1.378 ± 0.008 |
5.468 ± 0.014 | 6.055 ± 0.018 |
3.855 ± 0.012 | 6.917 ± 0.018 |
2.499 ± 0.009 | 4.957 ± 0.014 |
4.855 ± 0.016 | 8.829 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.202 ± 0.007 | 3.706 ± 0.01 |
6.21 ± 0.026 | 3.923 ± 0.013 |
6.157 ± 0.017 | 8.078 ± 0.019 |
5.817 ± 0.013 | 6.018 ± 0.013 |
1.594 ± 0.007 | 2.834 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
