
Muriicola sp. MMS17-SY002
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Muriicola
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
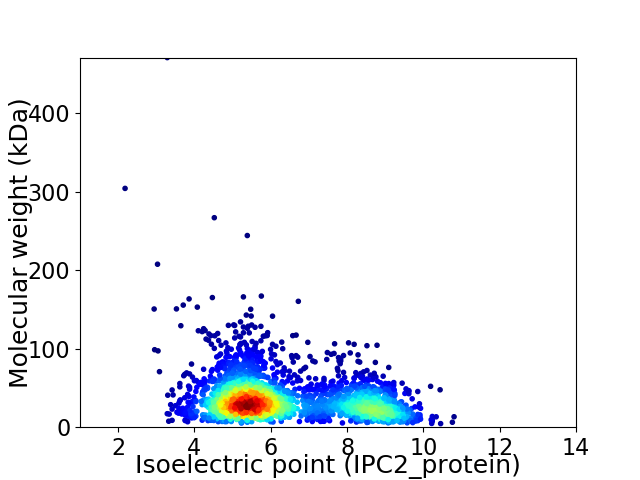
Virtual 2D-PAGE plot for 2593 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A411EC01|A0A411EC01_9FLAO Carbohydrate kinase OS=Muriicola sp. MMS17-SY002 OX=2507538 GN=EQY75_11330 PE=4 SV=1
MM1 pKa = 7.29IRR3 pKa = 11.84KK4 pKa = 8.94LLVLCLLAGSFSYY17 pKa = 11.0AQFNQNAPWLAGPDD31 pKa = 3.64EE32 pKa = 4.9SEE34 pKa = 4.36LRR36 pKa = 11.84TPSTVQPLSIFEE48 pKa = 4.12ISASAEE54 pKa = 4.08AYY56 pKa = 7.68WKK58 pKa = 11.16NKK60 pKa = 10.47DD61 pKa = 3.41KK62 pKa = 10.86DD63 pKa = 3.98AKK65 pKa = 10.54GSGYY69 pKa = 10.67KK70 pKa = 9.83PFKK73 pKa = 9.96RR74 pKa = 11.84WEE76 pKa = 4.45DD77 pKa = 3.55YY78 pKa = 10.03WMHH81 pKa = 6.67FVDD84 pKa = 3.89QQGYY88 pKa = 9.67LPTPKK93 pKa = 10.06EE94 pKa = 3.83LWEE97 pKa = 4.0TWEE100 pKa = 4.12NKK102 pKa = 7.07QNRR105 pKa = 11.84IGMTTNPVSAWSSVGPEE122 pKa = 3.98TVGVFSGRR130 pKa = 11.84LPGTGRR136 pKa = 11.84TNAIEE141 pKa = 4.08VDD143 pKa = 3.83PNDD146 pKa = 3.83PNTWYY151 pKa = 10.7VGAPAGGIWKK161 pKa = 7.23TTDD164 pKa = 4.52AGNTWTNLFDD174 pKa = 5.48DD175 pKa = 5.51FPQIGVSSIAIDD187 pKa = 4.03PNDD190 pKa = 3.74SNIIYY195 pKa = 10.22IATGDD200 pKa = 3.81DD201 pKa = 3.75DD202 pKa = 5.65AADD205 pKa = 3.96SYY207 pKa = 11.81SVGVFKK213 pKa = 11.17SLDD216 pKa = 3.43AGATWQEE223 pKa = 4.04TGLNPSTSNFSTLMTVILIDD243 pKa = 3.75PTDD246 pKa = 4.3SNVLWVATNTGLYY259 pKa = 10.14KK260 pKa = 10.81SIDD263 pKa = 3.85AGDD266 pKa = 3.35TWNIKK271 pKa = 9.11QGGYY275 pKa = 9.75IADD278 pKa = 4.41LKK280 pKa = 10.89LKK282 pKa = 10.34PGDD285 pKa = 3.8ANTVYY290 pKa = 10.74AIVGRR295 pKa = 11.84YY296 pKa = 9.38LGGSGNQVTFYY307 pKa = 11.0KK308 pKa = 9.99STDD311 pKa = 3.3GGDD314 pKa = 3.74TPFVALDD321 pKa = 4.14DD322 pKa = 4.72PLLPTSSGRR331 pKa = 11.84ALLGVTPADD340 pKa = 3.64PEE342 pKa = 4.03VLYY345 pKa = 10.56ILTANTGSSNFTFQGLYY362 pKa = 10.45KK363 pKa = 9.87STDD366 pKa = 3.06SGEE369 pKa = 4.42TFTEE373 pKa = 4.5SPNTTNIMEE382 pKa = 4.45SSQAWFDD389 pKa = 3.57LAFEE393 pKa = 4.23VSPTNANEE401 pKa = 4.53LYY403 pKa = 9.85MGCLNIWKK411 pKa = 9.71SVNGGNSFTRR421 pKa = 11.84LNQWFTNNAAYY432 pKa = 8.56THH434 pKa = 6.91ADD436 pKa = 2.43IHH438 pKa = 5.86TIKK441 pKa = 10.41IFNDD445 pKa = 3.13QVFACTDD452 pKa = 3.16GGIYY456 pKa = 10.35RR457 pKa = 11.84STNGGGSFTDD467 pKa = 3.92YY468 pKa = 10.18TSGMAVGQFYY478 pKa = 10.65RR479 pKa = 11.84LSVSPEE485 pKa = 3.79NSSKK489 pKa = 10.58MIGGLQDD496 pKa = 3.0NGGQILQNGVWNNYY510 pKa = 9.28HH511 pKa = 6.7GGDD514 pKa = 4.03GMDD517 pKa = 4.62NVIDD521 pKa = 4.25PNNDD525 pKa = 2.67NLVYY529 pKa = 10.78GFTQFGGSLNISSNSGQSIGFVGPPRR555 pKa = 11.84DD556 pKa = 3.89DD557 pKa = 3.57QNNTIQGNWITPLAISSTGEE577 pKa = 3.99VYY579 pKa = 10.83SGFDD583 pKa = 3.05AVYY586 pKa = 10.56KK587 pKa = 10.5LVGSAWEE594 pKa = 3.9KK595 pKa = 9.28WSNDD599 pKa = 3.17FGDD602 pKa = 4.37GNIDD606 pKa = 4.65DD607 pKa = 5.07IEE609 pKa = 4.29VDD611 pKa = 3.46PTDD614 pKa = 4.02PNVIYY619 pKa = 10.19AAEE622 pKa = 4.15GNIIYY627 pKa = 10.25RR628 pKa = 11.84SEE630 pKa = 4.7DD631 pKa = 2.81GGQNFTTFYY640 pKa = 10.78LADD643 pKa = 4.11ADD645 pKa = 3.86ISDD648 pKa = 3.78MTVNSNDD655 pKa = 2.98GSFVYY660 pKa = 10.61YY661 pKa = 8.6VTSRR665 pKa = 11.84RR666 pKa = 11.84VGISQANQPTQRR678 pKa = 11.84EE679 pKa = 4.35IYY681 pKa = 9.57KK682 pKa = 10.35IPVNEE687 pKa = 4.05NGEE690 pKa = 4.17AGLEE694 pKa = 3.82VDD696 pKa = 3.09ITYY699 pKa = 10.09NIPADD704 pKa = 3.37QAFFSIVHH712 pKa = 5.81QGRR715 pKa = 11.84HH716 pKa = 5.11TDD718 pKa = 3.01NPLYY722 pKa = 10.74VGTSLGVYY730 pKa = 8.72RR731 pKa = 11.84TDD733 pKa = 3.46DD734 pKa = 3.62TLSEE738 pKa = 4.05WEE740 pKa = 5.29DD741 pKa = 4.03YY742 pKa = 9.28FTDD745 pKa = 4.14LPSVAVSDD753 pKa = 5.01LDD755 pKa = 3.78ISLDD759 pKa = 3.77DD760 pKa = 4.22EE761 pKa = 5.83LITASTYY768 pKa = 10.82GRR770 pKa = 11.84GVWQSPIPIQVPDD783 pKa = 3.41NDD785 pKa = 3.46VRR787 pKa = 11.84LVSLSPANDD796 pKa = 3.18LVLCGEE802 pKa = 5.28IIPEE806 pKa = 3.55IVVEE810 pKa = 4.15NNGLNPITSVDD821 pKa = 3.15VSYY824 pKa = 10.5TIDD827 pKa = 3.67GGNPQNFNAPVNLNSEE843 pKa = 4.29EE844 pKa = 4.27TTTISLPSLNLTTIGQYY861 pKa = 8.14TLQVTVSITGDD872 pKa = 2.95AFADD876 pKa = 3.64NNEE879 pKa = 3.91ISHH882 pKa = 6.11SFFVNGFGVGDD893 pKa = 4.42EE894 pKa = 4.33INLFEE899 pKa = 4.54STADD903 pKa = 3.67NLNTYY908 pKa = 10.16NDD910 pKa = 3.92GGGAPLWEE918 pKa = 4.24RR919 pKa = 11.84GVPEE923 pKa = 4.92GSLLNQARR931 pKa = 11.84SGTQVYY937 pKa = 7.56GTNLDD942 pKa = 3.76GNHH945 pKa = 7.38PDD947 pKa = 3.06GTIAYY952 pKa = 8.17LISDD956 pKa = 4.79CYY958 pKa = 10.36EE959 pKa = 3.99LSSILAPVLKK969 pKa = 10.38FQMAYY974 pKa = 10.47DD975 pKa = 3.93LEE977 pKa = 4.68INFDD981 pKa = 3.48IVYY984 pKa = 10.24VEE986 pKa = 4.17YY987 pKa = 9.99STDD990 pKa = 5.27DD991 pKa = 3.52GANWQVLGSVNSQPNWYY1008 pKa = 10.43NSDD1011 pKa = 3.52RR1012 pKa = 11.84TNEE1015 pKa = 4.0SSGADD1020 pKa = 4.09DD1021 pKa = 5.58DD1022 pKa = 4.96CQNCPGAQWTGTEE1035 pKa = 3.98ATLTEE1040 pKa = 4.01YY1041 pKa = 10.78AYY1043 pKa = 10.83DD1044 pKa = 4.14FTANAALGEE1053 pKa = 4.12TDD1055 pKa = 4.03LTNEE1059 pKa = 3.73NNVIFRR1065 pKa = 11.84IVFHH1069 pKa = 7.1SDD1071 pKa = 2.71PSVNQEE1077 pKa = 3.51GAIIDD1082 pKa = 3.97DD1083 pKa = 3.99LVVEE1087 pKa = 5.15GFQDD1091 pKa = 5.88DD1092 pKa = 5.55DD1093 pKa = 6.19DD1094 pKa = 6.24DD1095 pKa = 6.88DD1096 pKa = 5.82NDD1098 pKa = 3.99GVLDD1102 pKa = 4.1VDD1104 pKa = 4.99DD1105 pKa = 4.74NCPLVGNADD1114 pKa = 3.74QLDD1117 pKa = 3.79TDD1119 pKa = 4.44GDD1121 pKa = 4.46GEE1123 pKa = 4.81GDD1125 pKa = 4.02ACDD1128 pKa = 4.15TDD1130 pKa = 4.59DD1131 pKa = 6.15DD1132 pKa = 5.01NDD1134 pKa = 5.23GIDD1137 pKa = 5.93DD1138 pKa = 4.56IDD1140 pKa = 4.84DD1141 pKa = 4.2NCPLIANADD1150 pKa = 3.69QADD1153 pKa = 3.64ADD1155 pKa = 4.1GDD1157 pKa = 4.53GIGDD1161 pKa = 3.83VCDD1164 pKa = 5.37DD1165 pKa = 4.52DD1166 pKa = 6.5ADD1168 pKa = 4.07NDD1170 pKa = 4.37GVPNAQDD1177 pKa = 3.73LCDD1180 pKa = 3.85NTPPGAVVDD1189 pKa = 4.05VTGCEE1194 pKa = 4.01IFTLPSNNFSLLAVGEE1210 pKa = 4.62TCTSNNDD1217 pKa = 2.81GSIVVNATEE1226 pKa = 4.16ALNYY1230 pKa = 8.63TAILSGGGLNQSQSFSDD1247 pKa = 3.26TFTFEE1252 pKa = 4.06NLASGSYY1259 pKa = 9.78DD1260 pKa = 3.13LCITVEE1266 pKa = 4.14GQPGYY1271 pKa = 8.65EE1272 pKa = 3.81VCYY1275 pKa = 9.94SVSISEE1281 pKa = 4.5PEE1283 pKa = 3.98DD1284 pKa = 3.64LSVSSKK1290 pKa = 10.35IDD1292 pKa = 3.3SFDD1295 pKa = 3.6NKK1297 pKa = 9.1VTLDD1301 pKa = 3.52LSGGKK1306 pKa = 9.8SYY1308 pKa = 10.35TINLNGEE1315 pKa = 3.89IFRR1318 pKa = 11.84TSEE1321 pKa = 4.17RR1322 pKa = 11.84EE1323 pKa = 3.37ITLPLSQVEE1332 pKa = 4.16NTLTVRR1338 pKa = 11.84TDD1340 pKa = 3.26KK1341 pKa = 11.08DD1342 pKa = 3.7CQGVYY1347 pKa = 10.44SEE1349 pKa = 5.09TIMLSSEE1356 pKa = 3.75LLIYY1360 pKa = 8.91PNPIASGDD1368 pKa = 3.73LNIYY1372 pKa = 9.82LGNNSSSQVEE1382 pKa = 4.31VALFDD1387 pKa = 4.64LNGRR1391 pKa = 11.84TVFRR1395 pKa = 11.84KK1396 pKa = 9.66EE1397 pKa = 3.57YY1398 pKa = 9.94NVQNNEE1404 pKa = 3.29IKK1406 pKa = 10.88FNVDD1410 pKa = 2.92ALSKK1414 pKa = 10.4GIYY1417 pKa = 8.29MLNIKK1422 pKa = 9.21TSSSLMNYY1430 pKa = 9.94KK1431 pKa = 10.22IIRR1434 pKa = 11.84KK1435 pKa = 9.01
MM1 pKa = 7.29IRR3 pKa = 11.84KK4 pKa = 8.94LLVLCLLAGSFSYY17 pKa = 11.0AQFNQNAPWLAGPDD31 pKa = 3.64EE32 pKa = 4.9SEE34 pKa = 4.36LRR36 pKa = 11.84TPSTVQPLSIFEE48 pKa = 4.12ISASAEE54 pKa = 4.08AYY56 pKa = 7.68WKK58 pKa = 11.16NKK60 pKa = 10.47DD61 pKa = 3.41KK62 pKa = 10.86DD63 pKa = 3.98AKK65 pKa = 10.54GSGYY69 pKa = 10.67KK70 pKa = 9.83PFKK73 pKa = 9.96RR74 pKa = 11.84WEE76 pKa = 4.45DD77 pKa = 3.55YY78 pKa = 10.03WMHH81 pKa = 6.67FVDD84 pKa = 3.89QQGYY88 pKa = 9.67LPTPKK93 pKa = 10.06EE94 pKa = 3.83LWEE97 pKa = 4.0TWEE100 pKa = 4.12NKK102 pKa = 7.07QNRR105 pKa = 11.84IGMTTNPVSAWSSVGPEE122 pKa = 3.98TVGVFSGRR130 pKa = 11.84LPGTGRR136 pKa = 11.84TNAIEE141 pKa = 4.08VDD143 pKa = 3.83PNDD146 pKa = 3.83PNTWYY151 pKa = 10.7VGAPAGGIWKK161 pKa = 7.23TTDD164 pKa = 4.52AGNTWTNLFDD174 pKa = 5.48DD175 pKa = 5.51FPQIGVSSIAIDD187 pKa = 4.03PNDD190 pKa = 3.74SNIIYY195 pKa = 10.22IATGDD200 pKa = 3.81DD201 pKa = 3.75DD202 pKa = 5.65AADD205 pKa = 3.96SYY207 pKa = 11.81SVGVFKK213 pKa = 11.17SLDD216 pKa = 3.43AGATWQEE223 pKa = 4.04TGLNPSTSNFSTLMTVILIDD243 pKa = 3.75PTDD246 pKa = 4.3SNVLWVATNTGLYY259 pKa = 10.14KK260 pKa = 10.81SIDD263 pKa = 3.85AGDD266 pKa = 3.35TWNIKK271 pKa = 9.11QGGYY275 pKa = 9.75IADD278 pKa = 4.41LKK280 pKa = 10.89LKK282 pKa = 10.34PGDD285 pKa = 3.8ANTVYY290 pKa = 10.74AIVGRR295 pKa = 11.84YY296 pKa = 9.38LGGSGNQVTFYY307 pKa = 11.0KK308 pKa = 9.99STDD311 pKa = 3.3GGDD314 pKa = 3.74TPFVALDD321 pKa = 4.14DD322 pKa = 4.72PLLPTSSGRR331 pKa = 11.84ALLGVTPADD340 pKa = 3.64PEE342 pKa = 4.03VLYY345 pKa = 10.56ILTANTGSSNFTFQGLYY362 pKa = 10.45KK363 pKa = 9.87STDD366 pKa = 3.06SGEE369 pKa = 4.42TFTEE373 pKa = 4.5SPNTTNIMEE382 pKa = 4.45SSQAWFDD389 pKa = 3.57LAFEE393 pKa = 4.23VSPTNANEE401 pKa = 4.53LYY403 pKa = 9.85MGCLNIWKK411 pKa = 9.71SVNGGNSFTRR421 pKa = 11.84LNQWFTNNAAYY432 pKa = 8.56THH434 pKa = 6.91ADD436 pKa = 2.43IHH438 pKa = 5.86TIKK441 pKa = 10.41IFNDD445 pKa = 3.13QVFACTDD452 pKa = 3.16GGIYY456 pKa = 10.35RR457 pKa = 11.84STNGGGSFTDD467 pKa = 3.92YY468 pKa = 10.18TSGMAVGQFYY478 pKa = 10.65RR479 pKa = 11.84LSVSPEE485 pKa = 3.79NSSKK489 pKa = 10.58MIGGLQDD496 pKa = 3.0NGGQILQNGVWNNYY510 pKa = 9.28HH511 pKa = 6.7GGDD514 pKa = 4.03GMDD517 pKa = 4.62NVIDD521 pKa = 4.25PNNDD525 pKa = 2.67NLVYY529 pKa = 10.78GFTQFGGSLNISSNSGQSIGFVGPPRR555 pKa = 11.84DD556 pKa = 3.89DD557 pKa = 3.57QNNTIQGNWITPLAISSTGEE577 pKa = 3.99VYY579 pKa = 10.83SGFDD583 pKa = 3.05AVYY586 pKa = 10.56KK587 pKa = 10.5LVGSAWEE594 pKa = 3.9KK595 pKa = 9.28WSNDD599 pKa = 3.17FGDD602 pKa = 4.37GNIDD606 pKa = 4.65DD607 pKa = 5.07IEE609 pKa = 4.29VDD611 pKa = 3.46PTDD614 pKa = 4.02PNVIYY619 pKa = 10.19AAEE622 pKa = 4.15GNIIYY627 pKa = 10.25RR628 pKa = 11.84SEE630 pKa = 4.7DD631 pKa = 2.81GGQNFTTFYY640 pKa = 10.78LADD643 pKa = 4.11ADD645 pKa = 3.86ISDD648 pKa = 3.78MTVNSNDD655 pKa = 2.98GSFVYY660 pKa = 10.61YY661 pKa = 8.6VTSRR665 pKa = 11.84RR666 pKa = 11.84VGISQANQPTQRR678 pKa = 11.84EE679 pKa = 4.35IYY681 pKa = 9.57KK682 pKa = 10.35IPVNEE687 pKa = 4.05NGEE690 pKa = 4.17AGLEE694 pKa = 3.82VDD696 pKa = 3.09ITYY699 pKa = 10.09NIPADD704 pKa = 3.37QAFFSIVHH712 pKa = 5.81QGRR715 pKa = 11.84HH716 pKa = 5.11TDD718 pKa = 3.01NPLYY722 pKa = 10.74VGTSLGVYY730 pKa = 8.72RR731 pKa = 11.84TDD733 pKa = 3.46DD734 pKa = 3.62TLSEE738 pKa = 4.05WEE740 pKa = 5.29DD741 pKa = 4.03YY742 pKa = 9.28FTDD745 pKa = 4.14LPSVAVSDD753 pKa = 5.01LDD755 pKa = 3.78ISLDD759 pKa = 3.77DD760 pKa = 4.22EE761 pKa = 5.83LITASTYY768 pKa = 10.82GRR770 pKa = 11.84GVWQSPIPIQVPDD783 pKa = 3.41NDD785 pKa = 3.46VRR787 pKa = 11.84LVSLSPANDD796 pKa = 3.18LVLCGEE802 pKa = 5.28IIPEE806 pKa = 3.55IVVEE810 pKa = 4.15NNGLNPITSVDD821 pKa = 3.15VSYY824 pKa = 10.5TIDD827 pKa = 3.67GGNPQNFNAPVNLNSEE843 pKa = 4.29EE844 pKa = 4.27TTTISLPSLNLTTIGQYY861 pKa = 8.14TLQVTVSITGDD872 pKa = 2.95AFADD876 pKa = 3.64NNEE879 pKa = 3.91ISHH882 pKa = 6.11SFFVNGFGVGDD893 pKa = 4.42EE894 pKa = 4.33INLFEE899 pKa = 4.54STADD903 pKa = 3.67NLNTYY908 pKa = 10.16NDD910 pKa = 3.92GGGAPLWEE918 pKa = 4.24RR919 pKa = 11.84GVPEE923 pKa = 4.92GSLLNQARR931 pKa = 11.84SGTQVYY937 pKa = 7.56GTNLDD942 pKa = 3.76GNHH945 pKa = 7.38PDD947 pKa = 3.06GTIAYY952 pKa = 8.17LISDD956 pKa = 4.79CYY958 pKa = 10.36EE959 pKa = 3.99LSSILAPVLKK969 pKa = 10.38FQMAYY974 pKa = 10.47DD975 pKa = 3.93LEE977 pKa = 4.68INFDD981 pKa = 3.48IVYY984 pKa = 10.24VEE986 pKa = 4.17YY987 pKa = 9.99STDD990 pKa = 5.27DD991 pKa = 3.52GANWQVLGSVNSQPNWYY1008 pKa = 10.43NSDD1011 pKa = 3.52RR1012 pKa = 11.84TNEE1015 pKa = 4.0SSGADD1020 pKa = 4.09DD1021 pKa = 5.58DD1022 pKa = 4.96CQNCPGAQWTGTEE1035 pKa = 3.98ATLTEE1040 pKa = 4.01YY1041 pKa = 10.78AYY1043 pKa = 10.83DD1044 pKa = 4.14FTANAALGEE1053 pKa = 4.12TDD1055 pKa = 4.03LTNEE1059 pKa = 3.73NNVIFRR1065 pKa = 11.84IVFHH1069 pKa = 7.1SDD1071 pKa = 2.71PSVNQEE1077 pKa = 3.51GAIIDD1082 pKa = 3.97DD1083 pKa = 3.99LVVEE1087 pKa = 5.15GFQDD1091 pKa = 5.88DD1092 pKa = 5.55DD1093 pKa = 6.19DD1094 pKa = 6.24DD1095 pKa = 6.88DD1096 pKa = 5.82NDD1098 pKa = 3.99GVLDD1102 pKa = 4.1VDD1104 pKa = 4.99DD1105 pKa = 4.74NCPLVGNADD1114 pKa = 3.74QLDD1117 pKa = 3.79TDD1119 pKa = 4.44GDD1121 pKa = 4.46GEE1123 pKa = 4.81GDD1125 pKa = 4.02ACDD1128 pKa = 4.15TDD1130 pKa = 4.59DD1131 pKa = 6.15DD1132 pKa = 5.01NDD1134 pKa = 5.23GIDD1137 pKa = 5.93DD1138 pKa = 4.56IDD1140 pKa = 4.84DD1141 pKa = 4.2NCPLIANADD1150 pKa = 3.69QADD1153 pKa = 3.64ADD1155 pKa = 4.1GDD1157 pKa = 4.53GIGDD1161 pKa = 3.83VCDD1164 pKa = 5.37DD1165 pKa = 4.52DD1166 pKa = 6.5ADD1168 pKa = 4.07NDD1170 pKa = 4.37GVPNAQDD1177 pKa = 3.73LCDD1180 pKa = 3.85NTPPGAVVDD1189 pKa = 4.05VTGCEE1194 pKa = 4.01IFTLPSNNFSLLAVGEE1210 pKa = 4.62TCTSNNDD1217 pKa = 2.81GSIVVNATEE1226 pKa = 4.16ALNYY1230 pKa = 8.63TAILSGGGLNQSQSFSDD1247 pKa = 3.26TFTFEE1252 pKa = 4.06NLASGSYY1259 pKa = 9.78DD1260 pKa = 3.13LCITVEE1266 pKa = 4.14GQPGYY1271 pKa = 8.65EE1272 pKa = 3.81VCYY1275 pKa = 9.94SVSISEE1281 pKa = 4.5PEE1283 pKa = 3.98DD1284 pKa = 3.64LSVSSKK1290 pKa = 10.35IDD1292 pKa = 3.3SFDD1295 pKa = 3.6NKK1297 pKa = 9.1VTLDD1301 pKa = 3.52LSGGKK1306 pKa = 9.8SYY1308 pKa = 10.35TINLNGEE1315 pKa = 3.89IFRR1318 pKa = 11.84TSEE1321 pKa = 4.17RR1322 pKa = 11.84EE1323 pKa = 3.37ITLPLSQVEE1332 pKa = 4.16NTLTVRR1338 pKa = 11.84TDD1340 pKa = 3.26KK1341 pKa = 11.08DD1342 pKa = 3.7CQGVYY1347 pKa = 10.44SEE1349 pKa = 5.09TIMLSSEE1356 pKa = 3.75LLIYY1360 pKa = 8.91PNPIASGDD1368 pKa = 3.73LNIYY1372 pKa = 9.82LGNNSSSQVEE1382 pKa = 4.31VALFDD1387 pKa = 4.64LNGRR1391 pKa = 11.84TVFRR1395 pKa = 11.84KK1396 pKa = 9.66EE1397 pKa = 3.57YY1398 pKa = 9.94NVQNNEE1404 pKa = 3.29IKK1406 pKa = 10.88FNVDD1410 pKa = 2.92ALSKK1414 pKa = 10.4GIYY1417 pKa = 8.29MLNIKK1422 pKa = 9.21TSSSLMNYY1430 pKa = 9.94KK1431 pKa = 10.22IIRR1434 pKa = 11.84KK1435 pKa = 9.01
Molecular weight: 155.53 kDa
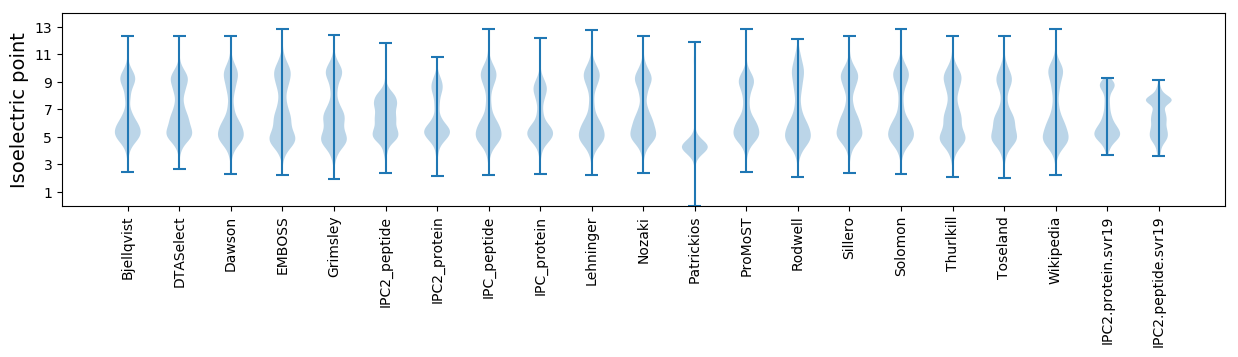
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A411ECB5|A0A411ECB5_9FLAO Class I SAM-dependent methyltransferase OS=Muriicola sp. MMS17-SY002 OX=2507538 GN=EQY75_13070 PE=4 SV=1
MM1 pKa = 7.87PKK3 pKa = 10.35NNSSRR8 pKa = 11.84SWIDD12 pKa = 2.81IVTEE16 pKa = 3.86LVEE19 pKa = 5.08LFFKK23 pKa = 10.88SSSRR27 pKa = 11.84SKK29 pKa = 10.72RR30 pKa = 11.84GRR32 pKa = 11.84TWHH35 pKa = 4.78QHH37 pKa = 3.25VVPYY41 pKa = 10.06EE42 pKa = 4.0NAWAVRR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 4.02GNKK53 pKa = 10.25RR54 pKa = 11.84ITSKK58 pKa = 10.32HH59 pKa = 5.07RR60 pKa = 11.84RR61 pKa = 11.84QDD63 pKa = 3.31TAINKK68 pKa = 9.21AKK70 pKa = 10.53RR71 pKa = 11.84LARR74 pKa = 11.84KK75 pKa = 9.28YY76 pKa = 9.39KK77 pKa = 10.16ADD79 pKa = 3.92VIIHH83 pKa = 6.54RR84 pKa = 11.84EE85 pKa = 3.94DD86 pKa = 3.32GSIRR90 pKa = 11.84DD91 pKa = 4.16RR92 pKa = 11.84INYY95 pKa = 8.03DD96 pKa = 2.82
MM1 pKa = 7.87PKK3 pKa = 10.35NNSSRR8 pKa = 11.84SWIDD12 pKa = 2.81IVTEE16 pKa = 3.86LVEE19 pKa = 5.08LFFKK23 pKa = 10.88SSSRR27 pKa = 11.84SKK29 pKa = 10.72RR30 pKa = 11.84GRR32 pKa = 11.84TWHH35 pKa = 4.78QHH37 pKa = 3.25VVPYY41 pKa = 10.06EE42 pKa = 4.0NAWAVRR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 4.02GNKK53 pKa = 10.25RR54 pKa = 11.84ITSKK58 pKa = 10.32HH59 pKa = 5.07RR60 pKa = 11.84RR61 pKa = 11.84QDD63 pKa = 3.31TAINKK68 pKa = 9.21AKK70 pKa = 10.53RR71 pKa = 11.84LARR74 pKa = 11.84KK75 pKa = 9.28YY76 pKa = 9.39KK77 pKa = 10.16ADD79 pKa = 3.92VIIHH83 pKa = 6.54RR84 pKa = 11.84EE85 pKa = 3.94DD86 pKa = 3.32GSIRR90 pKa = 11.84DD91 pKa = 4.16RR92 pKa = 11.84INYY95 pKa = 8.03DD96 pKa = 2.82
Molecular weight: 11.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
868456 |
38 |
4472 |
334.9 |
37.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.787 ± 0.042 | 0.726 ± 0.02 |
5.685 ± 0.062 | 7.013 ± 0.047 |
5.054 ± 0.042 | 6.895 ± 0.056 |
1.79 ± 0.026 | 7.352 ± 0.047 |
6.922 ± 0.07 | 9.775 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.366 ± 0.026 | 5.184 ± 0.042 |
3.743 ± 0.033 | 3.39 ± 0.032 |
4.117 ± 0.038 | 6.452 ± 0.037 |
5.377 ± 0.057 | 6.32 ± 0.038 |
1.131 ± 0.019 | 3.922 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
