
Candidatus Macondimonas diazotrophica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Candidatus Macondimonas
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
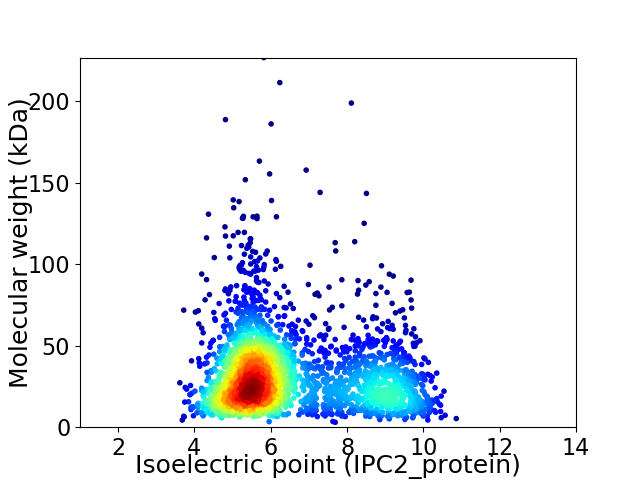
Virtual 2D-PAGE plot for 2722 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Z0F980|A0A4Z0F980_9GAMM NAD-dependent epimerase OS=Candidatus Macondimonas diazotrophica OX=2305248 GN=E4680_09365 PE=4 SV=1
MM1 pKa = 7.45SATSVVWAQSAPEE14 pKa = 3.9AASTIIAVQSFASPTLEE31 pKa = 4.06TPVGFGGSWGSVGVGAFGQTLADD54 pKa = 4.13DD55 pKa = 4.46EE56 pKa = 5.28FDD58 pKa = 3.42ASMGVAVGLGDD69 pKa = 3.73PDD71 pKa = 3.91RR72 pKa = 11.84YY73 pKa = 10.6LGAEE77 pKa = 4.09FGVGFSSLTGDD88 pKa = 3.32NSEE91 pKa = 5.36DD92 pKa = 3.56SFGDD96 pKa = 3.19SGGFSFKK103 pKa = 8.52VHH105 pKa = 6.81RR106 pKa = 11.84NLPGYY111 pKa = 9.45SAVSVGVNGFGVWGGAEE128 pKa = 4.22DD129 pKa = 4.95FNDD132 pKa = 3.53PSYY135 pKa = 10.89YY136 pKa = 10.24AAYY139 pKa = 10.69SKK141 pKa = 9.46MVPVGRR147 pKa = 11.84FASIVTIGAGTEE159 pKa = 4.16VFNDD163 pKa = 3.71PDD165 pKa = 4.51DD166 pKa = 4.91GGVNVFGSGALYY178 pKa = 7.55LTPQVALIGEE188 pKa = 4.26YY189 pKa = 9.54TGRR192 pKa = 11.84FVNAAVSVAPYY203 pKa = 9.95SWLPLTVTVGAVNVTEE219 pKa = 5.31RR220 pKa = 11.84FDD222 pKa = 4.69ADD224 pKa = 3.48TEE226 pKa = 4.35LAVSVGFGFNFTT238 pKa = 4.17
MM1 pKa = 7.45SATSVVWAQSAPEE14 pKa = 3.9AASTIIAVQSFASPTLEE31 pKa = 4.06TPVGFGGSWGSVGVGAFGQTLADD54 pKa = 4.13DD55 pKa = 4.46EE56 pKa = 5.28FDD58 pKa = 3.42ASMGVAVGLGDD69 pKa = 3.73PDD71 pKa = 3.91RR72 pKa = 11.84YY73 pKa = 10.6LGAEE77 pKa = 4.09FGVGFSSLTGDD88 pKa = 3.32NSEE91 pKa = 5.36DD92 pKa = 3.56SFGDD96 pKa = 3.19SGGFSFKK103 pKa = 8.52VHH105 pKa = 6.81RR106 pKa = 11.84NLPGYY111 pKa = 9.45SAVSVGVNGFGVWGGAEE128 pKa = 4.22DD129 pKa = 4.95FNDD132 pKa = 3.53PSYY135 pKa = 10.89YY136 pKa = 10.24AAYY139 pKa = 10.69SKK141 pKa = 9.46MVPVGRR147 pKa = 11.84FASIVTIGAGTEE159 pKa = 4.16VFNDD163 pKa = 3.71PDD165 pKa = 4.51DD166 pKa = 4.91GGVNVFGSGALYY178 pKa = 7.55LTPQVALIGEE188 pKa = 4.26YY189 pKa = 9.54TGRR192 pKa = 11.84FVNAAVSVAPYY203 pKa = 9.95SWLPLTVTVGAVNVTEE219 pKa = 5.31RR220 pKa = 11.84FDD222 pKa = 4.69ADD224 pKa = 3.48TEE226 pKa = 4.35LAVSVGFGFNFTT238 pKa = 4.17
Molecular weight: 24.41 kDa
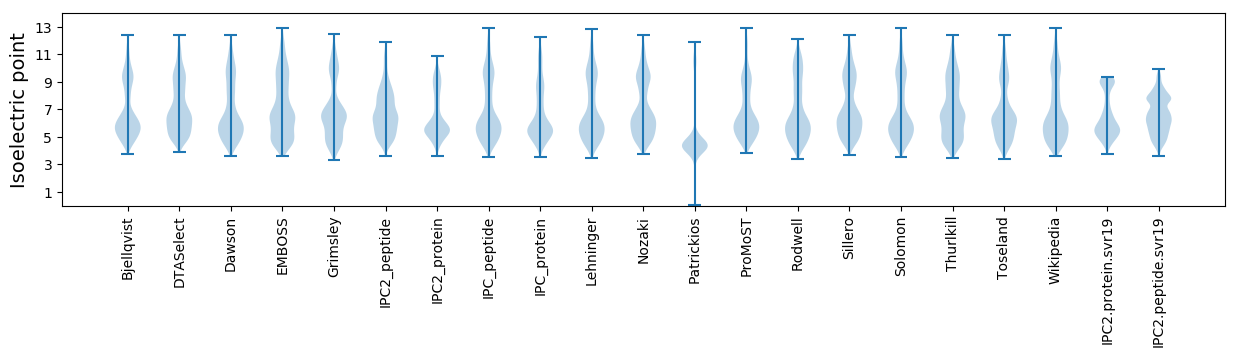
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Z0FBY2|A0A4Z0FBY2_9GAMM Uncharacterized protein OS=Candidatus Macondimonas diazotrophica OX=2305248 GN=E4680_05210 PE=4 SV=1
MM1 pKa = 6.96GTGVQYY7 pKa = 11.37VEE9 pKa = 4.83VSAEE13 pKa = 3.87RR14 pKa = 11.84AGQRR18 pKa = 11.84LDD20 pKa = 3.51NFLMGQLKK28 pKa = 9.79EE29 pKa = 4.01IPRR32 pKa = 11.84PQIYY36 pKa = 10.28RR37 pKa = 11.84ILRR40 pKa = 11.84KK41 pKa = 10.4GEE43 pKa = 3.56VRR45 pKa = 11.84VNGRR49 pKa = 11.84RR50 pKa = 11.84AKK52 pKa = 10.15PDD54 pKa = 3.31LRR56 pKa = 11.84IQAGDD61 pKa = 3.7RR62 pKa = 11.84VRR64 pKa = 11.84LPPLQRR70 pKa = 11.84APAAEE75 pKa = 4.23VAIPPASLIGEE86 pKa = 3.9IAGRR90 pKa = 11.84ILYY93 pKa = 10.1EE94 pKa = 4.5DD95 pKa = 4.87DD96 pKa = 3.8GLLVVNKK103 pKa = 9.35PAGLAVHH110 pKa = 7.22AGTGVAYY117 pKa = 10.24GLIDD121 pKa = 3.4VLRR124 pKa = 11.84ALRR127 pKa = 11.84PHH129 pKa = 6.56CPEE132 pKa = 4.24LNLVHH137 pKa = 7.07RR138 pKa = 11.84LDD140 pKa = 4.73RR141 pKa = 11.84GTSGCLVATKK151 pKa = 9.57TRR153 pKa = 11.84SDD155 pKa = 3.77LLTLNRR161 pKa = 11.84AFADD165 pKa = 4.01RR166 pKa = 11.84RR167 pKa = 11.84CEE169 pKa = 3.93KK170 pKa = 10.77VYY172 pKa = 10.79LALTQGRR179 pKa = 11.84WGRR182 pKa = 11.84GEE184 pKa = 3.95VVRR187 pKa = 11.84RR188 pKa = 11.84QALDD192 pKa = 3.03IDD194 pKa = 4.0HH195 pKa = 6.85RR196 pKa = 11.84QGGEE200 pKa = 3.53RR201 pKa = 11.84TVRR204 pKa = 11.84VDD206 pKa = 3.25TAHH209 pKa = 6.33GQTAVSRR216 pKa = 11.84FRR218 pKa = 11.84QVEE221 pKa = 4.18RR222 pKa = 11.84FDD224 pKa = 3.7GASLVRR230 pKa = 11.84VAIEE234 pKa = 3.64TGRR237 pKa = 11.84THH239 pKa = 7.18QIRR242 pKa = 11.84VHH244 pKa = 5.99AADD247 pKa = 4.72AGHH250 pKa = 7.33PLAGDD255 pKa = 3.33DD256 pKa = 3.96RR257 pKa = 11.84YY258 pKa = 11.32GDD260 pKa = 3.56RR261 pKa = 11.84VFNRR265 pKa = 11.84QMARR269 pKa = 11.84LGLRR273 pKa = 11.84RR274 pKa = 11.84LFLHH278 pKa = 6.93AAALSLPLDD287 pKa = 3.63GRR289 pKa = 11.84EE290 pKa = 3.97PLQVEE295 pKa = 4.46APLPADD301 pKa = 3.69LSGVLKK307 pKa = 10.72ALRR310 pKa = 11.84GG311 pKa = 3.63
MM1 pKa = 6.96GTGVQYY7 pKa = 11.37VEE9 pKa = 4.83VSAEE13 pKa = 3.87RR14 pKa = 11.84AGQRR18 pKa = 11.84LDD20 pKa = 3.51NFLMGQLKK28 pKa = 9.79EE29 pKa = 4.01IPRR32 pKa = 11.84PQIYY36 pKa = 10.28RR37 pKa = 11.84ILRR40 pKa = 11.84KK41 pKa = 10.4GEE43 pKa = 3.56VRR45 pKa = 11.84VNGRR49 pKa = 11.84RR50 pKa = 11.84AKK52 pKa = 10.15PDD54 pKa = 3.31LRR56 pKa = 11.84IQAGDD61 pKa = 3.7RR62 pKa = 11.84VRR64 pKa = 11.84LPPLQRR70 pKa = 11.84APAAEE75 pKa = 4.23VAIPPASLIGEE86 pKa = 3.9IAGRR90 pKa = 11.84ILYY93 pKa = 10.1EE94 pKa = 4.5DD95 pKa = 4.87DD96 pKa = 3.8GLLVVNKK103 pKa = 9.35PAGLAVHH110 pKa = 7.22AGTGVAYY117 pKa = 10.24GLIDD121 pKa = 3.4VLRR124 pKa = 11.84ALRR127 pKa = 11.84PHH129 pKa = 6.56CPEE132 pKa = 4.24LNLVHH137 pKa = 7.07RR138 pKa = 11.84LDD140 pKa = 4.73RR141 pKa = 11.84GTSGCLVATKK151 pKa = 9.57TRR153 pKa = 11.84SDD155 pKa = 3.77LLTLNRR161 pKa = 11.84AFADD165 pKa = 4.01RR166 pKa = 11.84RR167 pKa = 11.84CEE169 pKa = 3.93KK170 pKa = 10.77VYY172 pKa = 10.79LALTQGRR179 pKa = 11.84WGRR182 pKa = 11.84GEE184 pKa = 3.95VVRR187 pKa = 11.84RR188 pKa = 11.84QALDD192 pKa = 3.03IDD194 pKa = 4.0HH195 pKa = 6.85RR196 pKa = 11.84QGGEE200 pKa = 3.53RR201 pKa = 11.84TVRR204 pKa = 11.84VDD206 pKa = 3.25TAHH209 pKa = 6.33GQTAVSRR216 pKa = 11.84FRR218 pKa = 11.84QVEE221 pKa = 4.18RR222 pKa = 11.84FDD224 pKa = 3.7GASLVRR230 pKa = 11.84VAIEE234 pKa = 3.64TGRR237 pKa = 11.84THH239 pKa = 7.18QIRR242 pKa = 11.84VHH244 pKa = 5.99AADD247 pKa = 4.72AGHH250 pKa = 7.33PLAGDD255 pKa = 3.33DD256 pKa = 3.96RR257 pKa = 11.84YY258 pKa = 11.32GDD260 pKa = 3.56RR261 pKa = 11.84VFNRR265 pKa = 11.84QMARR269 pKa = 11.84LGLRR273 pKa = 11.84RR274 pKa = 11.84LFLHH278 pKa = 6.93AAALSLPLDD287 pKa = 3.63GRR289 pKa = 11.84EE290 pKa = 3.97PLQVEE295 pKa = 4.46APLPADD301 pKa = 3.69LSGVLKK307 pKa = 10.72ALRR310 pKa = 11.84GG311 pKa = 3.63
Molecular weight: 34.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
837426 |
24 |
2010 |
307.7 |
33.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.679 ± 0.07 | 0.999 ± 0.014 |
5.644 ± 0.037 | 5.728 ± 0.04 |
3.408 ± 0.028 | 8.237 ± 0.044 |
2.462 ± 0.028 | 4.836 ± 0.035 |
2.601 ± 0.047 | 10.996 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.387 ± 0.022 | 2.495 ± 0.03 |
5.452 ± 0.033 | 3.842 ± 0.03 |
7.776 ± 0.054 | 5.091 ± 0.031 |
5.117 ± 0.036 | 7.292 ± 0.043 |
1.518 ± 0.024 | 2.438 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
