
[Clostridium] hylemonae DSM 15053
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Lachnoclostridium; [Clostridium] hylemonae
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
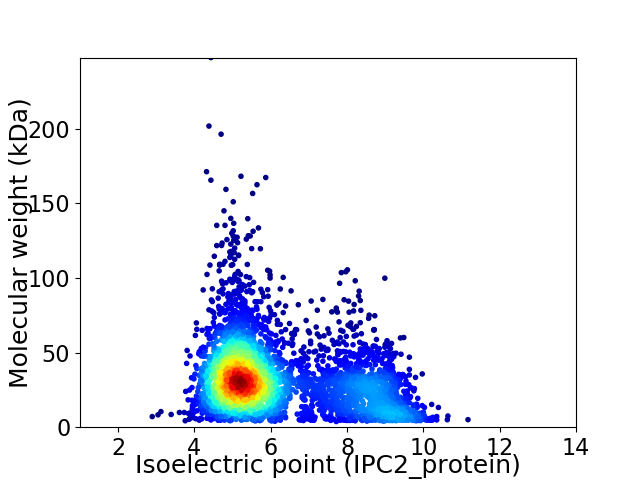
Virtual 2D-PAGE plot for 3971 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0BYY7|C0BYY7_9FIRM Uncharacterized protein OS=[Clostridium] hylemonae DSM 15053 OX=553973 GN=CLOHYLEM_05026 PE=4 SV=1
MM1 pKa = 6.99MKK3 pKa = 9.02KK4 pKa = 8.45TVALFMTVLMVFSLAACGSGSDD26 pKa = 4.49GKK28 pKa = 11.24GDD30 pKa = 3.7SKK32 pKa = 11.32KK33 pKa = 11.04EE34 pKa = 3.84EE35 pKa = 4.23DD36 pKa = 3.8EE37 pKa = 4.39KK38 pKa = 11.66GGDD41 pKa = 3.66SEE43 pKa = 5.18VVLEE47 pKa = 4.08FQQWFDD53 pKa = 3.43NEE55 pKa = 4.17MEE57 pKa = 4.11EE58 pKa = 6.03GYY60 pKa = 10.12LQSICDD66 pKa = 3.79DD67 pKa = 4.25FYY69 pKa = 11.38EE70 pKa = 4.23EE71 pKa = 3.97TGIKK75 pKa = 9.68IKK77 pKa = 10.76LLSNPYY83 pKa = 10.76ADD85 pKa = 3.69TKK87 pKa = 9.89TQLEE91 pKa = 4.1ASAAAGTMADD101 pKa = 3.68IVALDD106 pKa = 3.81GGWIYY111 pKa = 11.04DD112 pKa = 3.71YY113 pKa = 11.56AKK115 pKa = 10.57QGLLAKK121 pKa = 10.44LDD123 pKa = 3.7GLYY126 pKa = 10.77EE127 pKa = 4.11DD128 pKa = 4.43TGFDD132 pKa = 3.3AKK134 pKa = 10.77SVAEE138 pKa = 4.12GTKK141 pKa = 10.05VDD143 pKa = 3.55KK144 pKa = 10.88ALYY147 pKa = 8.77AQPIVNFPSMMAVNLDD163 pKa = 3.81LLKK166 pKa = 10.98SAGVDD171 pKa = 3.38TLPEE175 pKa = 3.97TWSEE179 pKa = 3.92FQEE182 pKa = 4.53ACEE185 pKa = 4.22KK186 pKa = 10.18VTDD189 pKa = 4.29SGDD192 pKa = 3.35NVYY195 pKa = 10.67GFCSNMSTDD204 pKa = 3.1NATSVEE210 pKa = 4.39YY211 pKa = 9.38LTAFAWNSGCSILTDD226 pKa = 3.8DD227 pKa = 5.14GKK229 pKa = 10.16PFIADD234 pKa = 3.5NEE236 pKa = 4.38VLADD240 pKa = 3.4TCEE243 pKa = 3.98FLKK246 pKa = 11.14GLIDD250 pKa = 4.52AKK252 pKa = 9.89VTSPGIYY259 pKa = 10.11TMTDD263 pKa = 2.72ADD265 pKa = 3.76KK266 pKa = 11.62VEE268 pKa = 4.26EE269 pKa = 4.19FTNGRR274 pKa = 11.84VAFIADD280 pKa = 3.84SVAHH284 pKa = 5.69MSNIRR289 pKa = 11.84EE290 pKa = 4.08QAPDD294 pKa = 3.31MNFTYY299 pKa = 10.65MNMPHH304 pKa = 7.0KK305 pKa = 10.28DD306 pKa = 3.61DD307 pKa = 4.11YY308 pKa = 12.01SGDD311 pKa = 3.46SYY313 pKa = 11.96VRR315 pKa = 11.84VNNWAVGIGEE325 pKa = 3.94NCKK328 pKa = 10.66YY329 pKa = 10.51KK330 pKa = 10.94EE331 pKa = 3.88EE332 pKa = 3.88AAKK335 pKa = 10.41FIEE338 pKa = 4.35YY339 pKa = 10.52LLSPEE344 pKa = 4.27VNADD348 pKa = 3.59LCIHH352 pKa = 6.84ANAFPANSEE361 pKa = 4.36AEE363 pKa = 4.06PQYY366 pKa = 11.35EE367 pKa = 4.28DD368 pKa = 5.35DD369 pKa = 4.21SDD371 pKa = 3.99AFKK374 pKa = 10.83AIYY377 pKa = 9.37DD378 pKa = 3.56VYY380 pKa = 10.47MNSNGVAEE388 pKa = 4.77YY389 pKa = 11.0YY390 pKa = 11.23SMPTAEE396 pKa = 4.86ALMSAFIDD404 pKa = 3.97GLVMYY409 pKa = 10.23FDD411 pKa = 5.03GDD413 pKa = 3.77YY414 pKa = 11.54DD415 pKa = 4.27DD416 pKa = 5.27VNDD419 pKa = 4.4MLSEE423 pKa = 4.15VQTQFDD429 pKa = 3.46AAYY432 pKa = 8.72EE433 pKa = 4.06
MM1 pKa = 6.99MKK3 pKa = 9.02KK4 pKa = 8.45TVALFMTVLMVFSLAACGSGSDD26 pKa = 4.49GKK28 pKa = 11.24GDD30 pKa = 3.7SKK32 pKa = 11.32KK33 pKa = 11.04EE34 pKa = 3.84EE35 pKa = 4.23DD36 pKa = 3.8EE37 pKa = 4.39KK38 pKa = 11.66GGDD41 pKa = 3.66SEE43 pKa = 5.18VVLEE47 pKa = 4.08FQQWFDD53 pKa = 3.43NEE55 pKa = 4.17MEE57 pKa = 4.11EE58 pKa = 6.03GYY60 pKa = 10.12LQSICDD66 pKa = 3.79DD67 pKa = 4.25FYY69 pKa = 11.38EE70 pKa = 4.23EE71 pKa = 3.97TGIKK75 pKa = 9.68IKK77 pKa = 10.76LLSNPYY83 pKa = 10.76ADD85 pKa = 3.69TKK87 pKa = 9.89TQLEE91 pKa = 4.1ASAAAGTMADD101 pKa = 3.68IVALDD106 pKa = 3.81GGWIYY111 pKa = 11.04DD112 pKa = 3.71YY113 pKa = 11.56AKK115 pKa = 10.57QGLLAKK121 pKa = 10.44LDD123 pKa = 3.7GLYY126 pKa = 10.77EE127 pKa = 4.11DD128 pKa = 4.43TGFDD132 pKa = 3.3AKK134 pKa = 10.77SVAEE138 pKa = 4.12GTKK141 pKa = 10.05VDD143 pKa = 3.55KK144 pKa = 10.88ALYY147 pKa = 8.77AQPIVNFPSMMAVNLDD163 pKa = 3.81LLKK166 pKa = 10.98SAGVDD171 pKa = 3.38TLPEE175 pKa = 3.97TWSEE179 pKa = 3.92FQEE182 pKa = 4.53ACEE185 pKa = 4.22KK186 pKa = 10.18VTDD189 pKa = 4.29SGDD192 pKa = 3.35NVYY195 pKa = 10.67GFCSNMSTDD204 pKa = 3.1NATSVEE210 pKa = 4.39YY211 pKa = 9.38LTAFAWNSGCSILTDD226 pKa = 3.8DD227 pKa = 5.14GKK229 pKa = 10.16PFIADD234 pKa = 3.5NEE236 pKa = 4.38VLADD240 pKa = 3.4TCEE243 pKa = 3.98FLKK246 pKa = 11.14GLIDD250 pKa = 4.52AKK252 pKa = 9.89VTSPGIYY259 pKa = 10.11TMTDD263 pKa = 2.72ADD265 pKa = 3.76KK266 pKa = 11.62VEE268 pKa = 4.26EE269 pKa = 4.19FTNGRR274 pKa = 11.84VAFIADD280 pKa = 3.84SVAHH284 pKa = 5.69MSNIRR289 pKa = 11.84EE290 pKa = 4.08QAPDD294 pKa = 3.31MNFTYY299 pKa = 10.65MNMPHH304 pKa = 7.0KK305 pKa = 10.28DD306 pKa = 3.61DD307 pKa = 4.11YY308 pKa = 12.01SGDD311 pKa = 3.46SYY313 pKa = 11.96VRR315 pKa = 11.84VNNWAVGIGEE325 pKa = 3.94NCKK328 pKa = 10.66YY329 pKa = 10.51KK330 pKa = 10.94EE331 pKa = 3.88EE332 pKa = 3.88AAKK335 pKa = 10.41FIEE338 pKa = 4.35YY339 pKa = 10.52LLSPEE344 pKa = 4.27VNADD348 pKa = 3.59LCIHH352 pKa = 6.84ANAFPANSEE361 pKa = 4.36AEE363 pKa = 4.06PQYY366 pKa = 11.35EE367 pKa = 4.28DD368 pKa = 5.35DD369 pKa = 4.21SDD371 pKa = 3.99AFKK374 pKa = 10.83AIYY377 pKa = 9.37DD378 pKa = 3.56VYY380 pKa = 10.47MNSNGVAEE388 pKa = 4.77YY389 pKa = 11.0YY390 pKa = 11.23SMPTAEE396 pKa = 4.86ALMSAFIDD404 pKa = 3.97GLVMYY409 pKa = 10.23FDD411 pKa = 5.03GDD413 pKa = 3.77YY414 pKa = 11.54DD415 pKa = 4.27DD416 pKa = 5.27VNDD419 pKa = 4.4MLSEE423 pKa = 4.15VQTQFDD429 pKa = 3.46AAYY432 pKa = 8.72EE433 pKa = 4.06
Molecular weight: 47.71 kDa
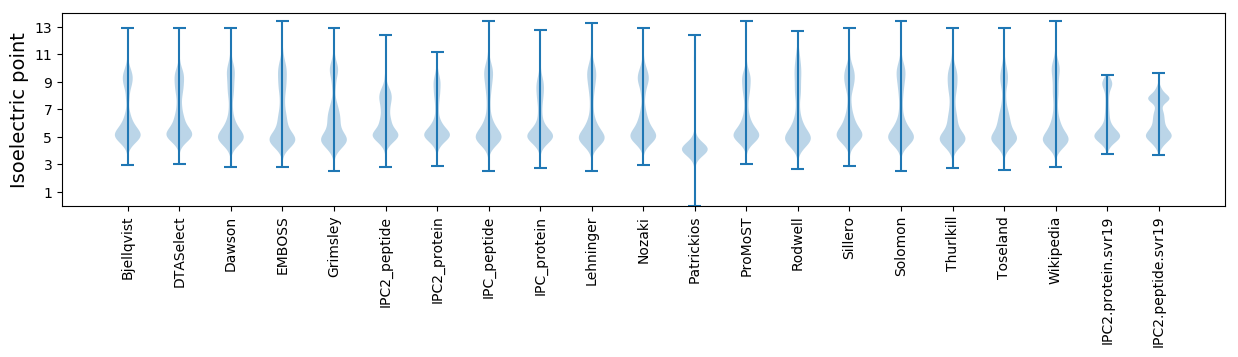
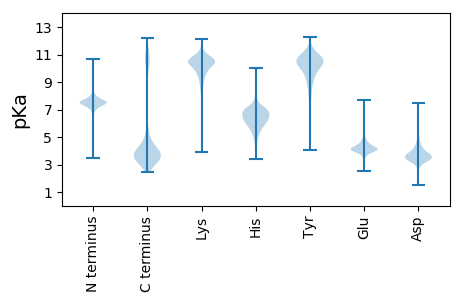
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0C332|C0C332_9FIRM 1-deoxy-D-xylulose 5-phosphate reductoisomerase OS=[Clostridium] hylemonae DSM 15053 OX=553973 GN=dxr PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.2VHH16 pKa = 5.65GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.72VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.92RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.2VHH16 pKa = 5.65GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.72VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.92RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1201420 |
35 |
2254 |
302.5 |
33.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.087 ± 0.041 | 1.576 ± 0.018 |
5.585 ± 0.036 | 7.678 ± 0.049 |
3.926 ± 0.03 | 7.785 ± 0.038 |
1.707 ± 0.018 | 7.08 ± 0.041 |
6.396 ± 0.03 | 8.908 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.221 ± 0.022 | 3.8 ± 0.024 |
3.388 ± 0.027 | 2.98 ± 0.021 |
4.882 ± 0.038 | 5.659 ± 0.026 |
5.325 ± 0.033 | 7.159 ± 0.032 |
0.859 ± 0.013 | 4.0 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
