
Tuber aestivum mitovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.79
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
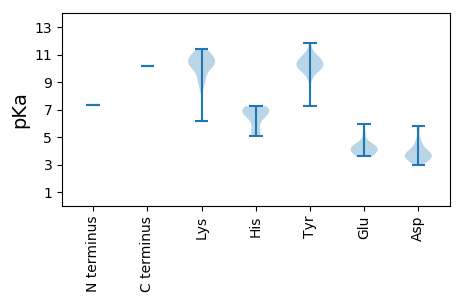
Protein with the lowest isoelectric point:
>tr|F6LFY6|F6LFY6_9VIRU RNA-dependent RNA polymerase OS=Tuber aestivum mitovirus OX=1037523 PE=4 SV=1
MM1 pKa = 7.36MNPFIRR7 pKa = 11.84LGSLLMGKK15 pKa = 6.16TTKK18 pKa = 10.47EE19 pKa = 4.05LIIGLYY25 pKa = 10.26SCVQTLHH32 pKa = 7.06KK33 pKa = 10.06IYY35 pKa = 10.37LSQGSRR41 pKa = 11.84GLAIRR46 pKa = 11.84LKK48 pKa = 10.79ASSTYY53 pKa = 11.03LMKK56 pKa = 10.5SWSGDD61 pKa = 3.37PLLDD65 pKa = 3.93CKK67 pKa = 11.2AFGPTMSLTPRR78 pKa = 11.84GIPIWIPSAWRR89 pKa = 11.84CQISSRR95 pKa = 11.84NLAIFRR101 pKa = 11.84VTMTLCNLYY110 pKa = 10.22RR111 pKa = 11.84VLPYY115 pKa = 10.07QGKK118 pKa = 9.84LKK120 pKa = 10.8LSTITDD126 pKa = 3.59DD127 pKa = 4.69RR128 pKa = 11.84KK129 pKa = 11.09GSTPDD134 pKa = 3.68LMFSFIKK141 pKa = 10.71DD142 pKa = 3.1KK143 pKa = 10.62FAPNITNRR151 pKa = 11.84VPYY154 pKa = 9.86PGIFSWDD161 pKa = 3.66PKK163 pKa = 10.32PLLTRR168 pKa = 11.84GPGAQKK174 pKa = 10.5EE175 pKa = 4.37KK176 pKa = 10.08NTMSGFGMALRR187 pKa = 11.84AIHH190 pKa = 5.34EE191 pKa = 4.43TGMLDD196 pKa = 3.51HH197 pKa = 6.92LKK199 pKa = 10.57AYY201 pKa = 10.49ALYY204 pKa = 10.01TGLNPQLEE212 pKa = 4.49SLKK215 pKa = 10.78VIWEE219 pKa = 4.43GTRR222 pKa = 11.84KK223 pKa = 9.49HH224 pKa = 6.48CSIGPIGKK232 pKa = 9.04LAFKK236 pKa = 10.41QEE238 pKa = 4.09PGKK241 pKa = 10.85VRR243 pKa = 11.84VFAMVDD249 pKa = 4.5CITQWFLHH257 pKa = 6.46PLHH260 pKa = 7.0KK261 pKa = 10.84YY262 pKa = 9.69LFSVLRR268 pKa = 11.84TTKK271 pKa = 10.5EE272 pKa = 3.65DD273 pKa = 3.2ATFDD277 pKa = 3.55QEE279 pKa = 4.6KK280 pKa = 10.45GINLVRR286 pKa = 11.84LALSKK291 pKa = 11.14KK292 pKa = 9.26LDD294 pKa = 3.41KK295 pKa = 11.2SVFSFDD301 pKa = 4.34LSAATDD307 pKa = 4.13RR308 pKa = 11.84LPMDD312 pKa = 3.3IQMVILNSLTPFWLAEE328 pKa = 3.75KK329 pKa = 10.26AVRR332 pKa = 11.84HH333 pKa = 5.04GVKK336 pKa = 10.43GGLGDD341 pKa = 3.64AWADD345 pKa = 3.57LLVDD349 pKa = 3.81RR350 pKa = 11.84DD351 pKa = 4.02YY352 pKa = 11.8YY353 pKa = 10.39LPRR356 pKa = 11.84WSGYY360 pKa = 10.65ARR362 pKa = 11.84DD363 pKa = 3.79SKK365 pKa = 11.13VRR367 pKa = 11.84YY368 pKa = 9.03AVGQPMGALSSWAMLALTHH387 pKa = 6.72HH388 pKa = 6.94MIVQFAAASVGVTGWFKK405 pKa = 11.06EE406 pKa = 3.79YY407 pKa = 9.9MVLGDD412 pKa = 5.78DD413 pKa = 3.29IVIYY417 pKa = 10.56NSEE420 pKa = 3.96VAKK423 pKa = 10.58AYY425 pKa = 7.24STLMGTLGVGISDD438 pKa = 3.93TKK440 pKa = 11.32SLTSKK445 pKa = 10.23IGVFEE450 pKa = 3.99FAKK453 pKa = 10.55RR454 pKa = 11.84LMDD457 pKa = 5.63LEE459 pKa = 5.37GPCQGLPLAEE469 pKa = 4.04FAAARR474 pKa = 11.84FNLSILFQSFRR485 pKa = 11.84SRR487 pKa = 11.84TLYY490 pKa = 10.25PKK492 pKa = 9.83ISTFMRR498 pKa = 11.84FLGFGYY504 pKa = 10.21KK505 pKa = 10.08VLGSLGMRR513 pKa = 11.84LGDD516 pKa = 3.25MRR518 pKa = 11.84KK519 pKa = 9.03RR520 pKa = 11.84SGYY523 pKa = 10.03FEE525 pKa = 4.19TVAYY529 pKa = 10.09SPLVTDD535 pKa = 4.13KK536 pKa = 11.25SVNKK540 pKa = 9.15WSEE543 pKa = 3.63FFKK546 pKa = 10.95RR547 pKa = 11.84FSTYY551 pKa = 10.99DD552 pKa = 3.68LYY554 pKa = 10.7PLVEE558 pKa = 4.11ACVYY562 pKa = 9.98KK563 pKa = 10.48AYY565 pKa = 10.71HH566 pKa = 7.23LIPALTKK573 pKa = 9.9FDD575 pKa = 3.9RR576 pKa = 11.84WNLWRR581 pKa = 11.84TLFPHH586 pKa = 7.01FDD588 pKa = 3.63DD589 pKa = 5.68KK590 pKa = 11.43KK591 pKa = 10.08ILLIPEE597 pKa = 3.75VAGRR601 pKa = 11.84HH602 pKa = 5.26LEE604 pKa = 4.12SLFTSMLDD612 pKa = 3.49NKK614 pKa = 8.67VAKK617 pKa = 10.24YY618 pKa = 10.22KK619 pKa = 10.4ADD621 pKa = 2.98HH622 pKa = 5.86SVYY625 pKa = 10.69VNRR628 pKa = 11.84LNRR631 pKa = 11.84AEE633 pKa = 4.32VPTNDD638 pKa = 3.2WEE640 pKa = 5.92AIDD643 pKa = 3.85TLLSIMEE650 pKa = 4.42TSPPTSDD657 pKa = 5.05LLQDD661 pKa = 3.59PDD663 pKa = 3.8MNEE666 pKa = 3.88LGFGLSVSNYY676 pKa = 10.82DD677 pKa = 3.89EE678 pKa = 4.99DD679 pKa = 4.6NKK681 pKa = 11.28SKK683 pKa = 9.13TCSLSLPYY691 pKa = 10.13TVFTKK696 pKa = 10.29IPQLLDD702 pKa = 4.64SIDD705 pKa = 3.9DD706 pKa = 3.6WVEE709 pKa = 3.61PIIPDD714 pKa = 3.25KK715 pKa = 11.01HH716 pKa = 6.99PEE718 pKa = 3.59VSADD722 pKa = 3.33VNRR725 pKa = 11.84PLIHH729 pKa = 6.83KK730 pKa = 8.61PGKK733 pKa = 10.42VFVKK737 pKa = 10.46GIATGKK743 pKa = 9.91NEE745 pKa = 3.99NNKK748 pKa = 9.43VEE750 pKa = 4.23ITKK753 pKa = 9.28MLQDD757 pKa = 4.09KK758 pKa = 10.71KK759 pKa = 10.17FLAYY763 pKa = 9.5IIEE766 pKa = 4.4SGKK769 pKa = 8.25ITIYY773 pKa = 10.87SPAFQAMIRR782 pKa = 11.84ASWAKK787 pKa = 9.89AQSGSKK793 pKa = 10.14
MM1 pKa = 7.36MNPFIRR7 pKa = 11.84LGSLLMGKK15 pKa = 6.16TTKK18 pKa = 10.47EE19 pKa = 4.05LIIGLYY25 pKa = 10.26SCVQTLHH32 pKa = 7.06KK33 pKa = 10.06IYY35 pKa = 10.37LSQGSRR41 pKa = 11.84GLAIRR46 pKa = 11.84LKK48 pKa = 10.79ASSTYY53 pKa = 11.03LMKK56 pKa = 10.5SWSGDD61 pKa = 3.37PLLDD65 pKa = 3.93CKK67 pKa = 11.2AFGPTMSLTPRR78 pKa = 11.84GIPIWIPSAWRR89 pKa = 11.84CQISSRR95 pKa = 11.84NLAIFRR101 pKa = 11.84VTMTLCNLYY110 pKa = 10.22RR111 pKa = 11.84VLPYY115 pKa = 10.07QGKK118 pKa = 9.84LKK120 pKa = 10.8LSTITDD126 pKa = 3.59DD127 pKa = 4.69RR128 pKa = 11.84KK129 pKa = 11.09GSTPDD134 pKa = 3.68LMFSFIKK141 pKa = 10.71DD142 pKa = 3.1KK143 pKa = 10.62FAPNITNRR151 pKa = 11.84VPYY154 pKa = 9.86PGIFSWDD161 pKa = 3.66PKK163 pKa = 10.32PLLTRR168 pKa = 11.84GPGAQKK174 pKa = 10.5EE175 pKa = 4.37KK176 pKa = 10.08NTMSGFGMALRR187 pKa = 11.84AIHH190 pKa = 5.34EE191 pKa = 4.43TGMLDD196 pKa = 3.51HH197 pKa = 6.92LKK199 pKa = 10.57AYY201 pKa = 10.49ALYY204 pKa = 10.01TGLNPQLEE212 pKa = 4.49SLKK215 pKa = 10.78VIWEE219 pKa = 4.43GTRR222 pKa = 11.84KK223 pKa = 9.49HH224 pKa = 6.48CSIGPIGKK232 pKa = 9.04LAFKK236 pKa = 10.41QEE238 pKa = 4.09PGKK241 pKa = 10.85VRR243 pKa = 11.84VFAMVDD249 pKa = 4.5CITQWFLHH257 pKa = 6.46PLHH260 pKa = 7.0KK261 pKa = 10.84YY262 pKa = 9.69LFSVLRR268 pKa = 11.84TTKK271 pKa = 10.5EE272 pKa = 3.65DD273 pKa = 3.2ATFDD277 pKa = 3.55QEE279 pKa = 4.6KK280 pKa = 10.45GINLVRR286 pKa = 11.84LALSKK291 pKa = 11.14KK292 pKa = 9.26LDD294 pKa = 3.41KK295 pKa = 11.2SVFSFDD301 pKa = 4.34LSAATDD307 pKa = 4.13RR308 pKa = 11.84LPMDD312 pKa = 3.3IQMVILNSLTPFWLAEE328 pKa = 3.75KK329 pKa = 10.26AVRR332 pKa = 11.84HH333 pKa = 5.04GVKK336 pKa = 10.43GGLGDD341 pKa = 3.64AWADD345 pKa = 3.57LLVDD349 pKa = 3.81RR350 pKa = 11.84DD351 pKa = 4.02YY352 pKa = 11.8YY353 pKa = 10.39LPRR356 pKa = 11.84WSGYY360 pKa = 10.65ARR362 pKa = 11.84DD363 pKa = 3.79SKK365 pKa = 11.13VRR367 pKa = 11.84YY368 pKa = 9.03AVGQPMGALSSWAMLALTHH387 pKa = 6.72HH388 pKa = 6.94MIVQFAAASVGVTGWFKK405 pKa = 11.06EE406 pKa = 3.79YY407 pKa = 9.9MVLGDD412 pKa = 5.78DD413 pKa = 3.29IVIYY417 pKa = 10.56NSEE420 pKa = 3.96VAKK423 pKa = 10.58AYY425 pKa = 7.24STLMGTLGVGISDD438 pKa = 3.93TKK440 pKa = 11.32SLTSKK445 pKa = 10.23IGVFEE450 pKa = 3.99FAKK453 pKa = 10.55RR454 pKa = 11.84LMDD457 pKa = 5.63LEE459 pKa = 5.37GPCQGLPLAEE469 pKa = 4.04FAAARR474 pKa = 11.84FNLSILFQSFRR485 pKa = 11.84SRR487 pKa = 11.84TLYY490 pKa = 10.25PKK492 pKa = 9.83ISTFMRR498 pKa = 11.84FLGFGYY504 pKa = 10.21KK505 pKa = 10.08VLGSLGMRR513 pKa = 11.84LGDD516 pKa = 3.25MRR518 pKa = 11.84KK519 pKa = 9.03RR520 pKa = 11.84SGYY523 pKa = 10.03FEE525 pKa = 4.19TVAYY529 pKa = 10.09SPLVTDD535 pKa = 4.13KK536 pKa = 11.25SVNKK540 pKa = 9.15WSEE543 pKa = 3.63FFKK546 pKa = 10.95RR547 pKa = 11.84FSTYY551 pKa = 10.99DD552 pKa = 3.68LYY554 pKa = 10.7PLVEE558 pKa = 4.11ACVYY562 pKa = 9.98KK563 pKa = 10.48AYY565 pKa = 10.71HH566 pKa = 7.23LIPALTKK573 pKa = 9.9FDD575 pKa = 3.9RR576 pKa = 11.84WNLWRR581 pKa = 11.84TLFPHH586 pKa = 7.01FDD588 pKa = 3.63DD589 pKa = 5.68KK590 pKa = 11.43KK591 pKa = 10.08ILLIPEE597 pKa = 3.75VAGRR601 pKa = 11.84HH602 pKa = 5.26LEE604 pKa = 4.12SLFTSMLDD612 pKa = 3.49NKK614 pKa = 8.67VAKK617 pKa = 10.24YY618 pKa = 10.22KK619 pKa = 10.4ADD621 pKa = 2.98HH622 pKa = 5.86SVYY625 pKa = 10.69VNRR628 pKa = 11.84LNRR631 pKa = 11.84AEE633 pKa = 4.32VPTNDD638 pKa = 3.2WEE640 pKa = 5.92AIDD643 pKa = 3.85TLLSIMEE650 pKa = 4.42TSPPTSDD657 pKa = 5.05LLQDD661 pKa = 3.59PDD663 pKa = 3.8MNEE666 pKa = 3.88LGFGLSVSNYY676 pKa = 10.82DD677 pKa = 3.89EE678 pKa = 4.99DD679 pKa = 4.6NKK681 pKa = 11.28SKK683 pKa = 9.13TCSLSLPYY691 pKa = 10.13TVFTKK696 pKa = 10.29IPQLLDD702 pKa = 4.64SIDD705 pKa = 3.9DD706 pKa = 3.6WVEE709 pKa = 3.61PIIPDD714 pKa = 3.25KK715 pKa = 11.01HH716 pKa = 6.99PEE718 pKa = 3.59VSADD722 pKa = 3.33VNRR725 pKa = 11.84PLIHH729 pKa = 6.83KK730 pKa = 8.61PGKK733 pKa = 10.42VFVKK737 pKa = 10.46GIATGKK743 pKa = 9.91NEE745 pKa = 3.99NNKK748 pKa = 9.43VEE750 pKa = 4.23ITKK753 pKa = 9.28MLQDD757 pKa = 4.09KK758 pKa = 10.71KK759 pKa = 10.17FLAYY763 pKa = 9.5IIEE766 pKa = 4.4SGKK769 pKa = 8.25ITIYY773 pKa = 10.87SPAFQAMIRR782 pKa = 11.84ASWAKK787 pKa = 9.89AQSGSKK793 pKa = 10.14
Molecular weight: 89.39 kDa
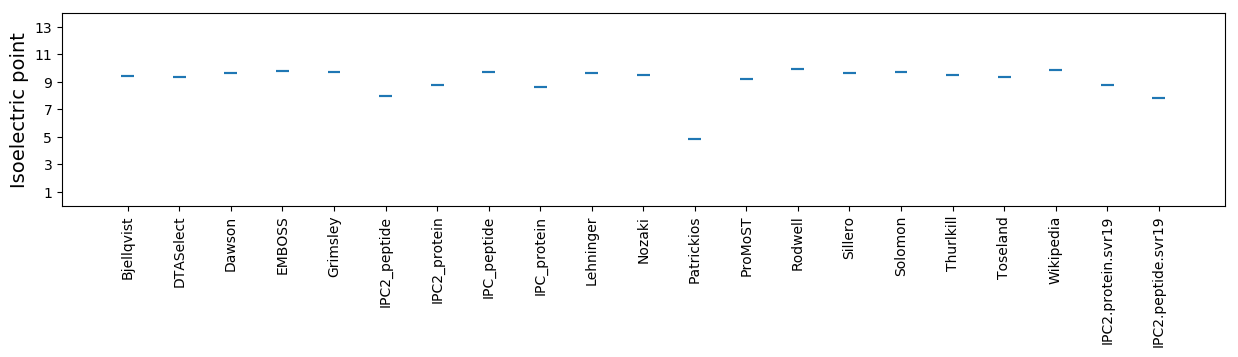
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F6LFY6|F6LFY6_9VIRU RNA-dependent RNA polymerase OS=Tuber aestivum mitovirus OX=1037523 PE=4 SV=1
MM1 pKa = 7.36MNPFIRR7 pKa = 11.84LGSLLMGKK15 pKa = 6.16TTKK18 pKa = 10.47EE19 pKa = 4.05LIIGLYY25 pKa = 10.26SCVQTLHH32 pKa = 7.06KK33 pKa = 10.06IYY35 pKa = 10.37LSQGSRR41 pKa = 11.84GLAIRR46 pKa = 11.84LKK48 pKa = 10.79ASSTYY53 pKa = 11.03LMKK56 pKa = 10.5SWSGDD61 pKa = 3.37PLLDD65 pKa = 3.93CKK67 pKa = 11.2AFGPTMSLTPRR78 pKa = 11.84GIPIWIPSAWRR89 pKa = 11.84CQISSRR95 pKa = 11.84NLAIFRR101 pKa = 11.84VTMTLCNLYY110 pKa = 10.22RR111 pKa = 11.84VLPYY115 pKa = 10.07QGKK118 pKa = 9.84LKK120 pKa = 10.8LSTITDD126 pKa = 3.59DD127 pKa = 4.69RR128 pKa = 11.84KK129 pKa = 11.09GSTPDD134 pKa = 3.68LMFSFIKK141 pKa = 10.71DD142 pKa = 3.1KK143 pKa = 10.62FAPNITNRR151 pKa = 11.84VPYY154 pKa = 9.86PGIFSWDD161 pKa = 3.66PKK163 pKa = 10.32PLLTRR168 pKa = 11.84GPGAQKK174 pKa = 10.5EE175 pKa = 4.37KK176 pKa = 10.08NTMSGFGMALRR187 pKa = 11.84AIHH190 pKa = 5.34EE191 pKa = 4.43TGMLDD196 pKa = 3.51HH197 pKa = 6.92LKK199 pKa = 10.57AYY201 pKa = 10.49ALYY204 pKa = 10.01TGLNPQLEE212 pKa = 4.49SLKK215 pKa = 10.78VIWEE219 pKa = 4.43GTRR222 pKa = 11.84KK223 pKa = 9.49HH224 pKa = 6.48CSIGPIGKK232 pKa = 9.04LAFKK236 pKa = 10.41QEE238 pKa = 4.09PGKK241 pKa = 10.85VRR243 pKa = 11.84VFAMVDD249 pKa = 4.5CITQWFLHH257 pKa = 6.46PLHH260 pKa = 7.0KK261 pKa = 10.84YY262 pKa = 9.69LFSVLRR268 pKa = 11.84TTKK271 pKa = 10.5EE272 pKa = 3.65DD273 pKa = 3.2ATFDD277 pKa = 3.55QEE279 pKa = 4.6KK280 pKa = 10.45GINLVRR286 pKa = 11.84LALSKK291 pKa = 11.14KK292 pKa = 9.26LDD294 pKa = 3.41KK295 pKa = 11.2SVFSFDD301 pKa = 4.34LSAATDD307 pKa = 4.13RR308 pKa = 11.84LPMDD312 pKa = 3.3IQMVILNSLTPFWLAEE328 pKa = 3.75KK329 pKa = 10.26AVRR332 pKa = 11.84HH333 pKa = 5.04GVKK336 pKa = 10.43GGLGDD341 pKa = 3.64AWADD345 pKa = 3.57LLVDD349 pKa = 3.81RR350 pKa = 11.84DD351 pKa = 4.02YY352 pKa = 11.8YY353 pKa = 10.39LPRR356 pKa = 11.84WSGYY360 pKa = 10.65ARR362 pKa = 11.84DD363 pKa = 3.79SKK365 pKa = 11.13VRR367 pKa = 11.84YY368 pKa = 9.03AVGQPMGALSSWAMLALTHH387 pKa = 6.72HH388 pKa = 6.94MIVQFAAASVGVTGWFKK405 pKa = 11.06EE406 pKa = 3.79YY407 pKa = 9.9MVLGDD412 pKa = 5.78DD413 pKa = 3.29IVIYY417 pKa = 10.56NSEE420 pKa = 3.96VAKK423 pKa = 10.58AYY425 pKa = 7.24STLMGTLGVGISDD438 pKa = 3.93TKK440 pKa = 11.32SLTSKK445 pKa = 10.23IGVFEE450 pKa = 3.99FAKK453 pKa = 10.55RR454 pKa = 11.84LMDD457 pKa = 5.63LEE459 pKa = 5.37GPCQGLPLAEE469 pKa = 4.04FAAARR474 pKa = 11.84FNLSILFQSFRR485 pKa = 11.84SRR487 pKa = 11.84TLYY490 pKa = 10.25PKK492 pKa = 9.83ISTFMRR498 pKa = 11.84FLGFGYY504 pKa = 10.21KK505 pKa = 10.08VLGSLGMRR513 pKa = 11.84LGDD516 pKa = 3.25MRR518 pKa = 11.84KK519 pKa = 9.03RR520 pKa = 11.84SGYY523 pKa = 10.03FEE525 pKa = 4.19TVAYY529 pKa = 10.09SPLVTDD535 pKa = 4.13KK536 pKa = 11.25SVNKK540 pKa = 9.15WSEE543 pKa = 3.63FFKK546 pKa = 10.95RR547 pKa = 11.84FSTYY551 pKa = 10.99DD552 pKa = 3.68LYY554 pKa = 10.7PLVEE558 pKa = 4.11ACVYY562 pKa = 9.98KK563 pKa = 10.48AYY565 pKa = 10.71HH566 pKa = 7.23LIPALTKK573 pKa = 9.9FDD575 pKa = 3.9RR576 pKa = 11.84WNLWRR581 pKa = 11.84TLFPHH586 pKa = 7.01FDD588 pKa = 3.63DD589 pKa = 5.68KK590 pKa = 11.43KK591 pKa = 10.08ILLIPEE597 pKa = 3.75VAGRR601 pKa = 11.84HH602 pKa = 5.26LEE604 pKa = 4.12SLFTSMLDD612 pKa = 3.49NKK614 pKa = 8.67VAKK617 pKa = 10.24YY618 pKa = 10.22KK619 pKa = 10.4ADD621 pKa = 2.98HH622 pKa = 5.86SVYY625 pKa = 10.69VNRR628 pKa = 11.84LNRR631 pKa = 11.84AEE633 pKa = 4.32VPTNDD638 pKa = 3.2WEE640 pKa = 5.92AIDD643 pKa = 3.85TLLSIMEE650 pKa = 4.42TSPPTSDD657 pKa = 5.05LLQDD661 pKa = 3.59PDD663 pKa = 3.8MNEE666 pKa = 3.88LGFGLSVSNYY676 pKa = 10.82DD677 pKa = 3.89EE678 pKa = 4.99DD679 pKa = 4.6NKK681 pKa = 11.28SKK683 pKa = 9.13TCSLSLPYY691 pKa = 10.13TVFTKK696 pKa = 10.29IPQLLDD702 pKa = 4.64SIDD705 pKa = 3.9DD706 pKa = 3.6WVEE709 pKa = 3.61PIIPDD714 pKa = 3.25KK715 pKa = 11.01HH716 pKa = 6.99PEE718 pKa = 3.59VSADD722 pKa = 3.33VNRR725 pKa = 11.84PLIHH729 pKa = 6.83KK730 pKa = 8.61PGKK733 pKa = 10.42VFVKK737 pKa = 10.46GIATGKK743 pKa = 9.91NEE745 pKa = 3.99NNKK748 pKa = 9.43VEE750 pKa = 4.23ITKK753 pKa = 9.28MLQDD757 pKa = 4.09KK758 pKa = 10.71KK759 pKa = 10.17FLAYY763 pKa = 9.5IIEE766 pKa = 4.4SGKK769 pKa = 8.25ITIYY773 pKa = 10.87SPAFQAMIRR782 pKa = 11.84ASWAKK787 pKa = 9.89AQSGSKK793 pKa = 10.14
MM1 pKa = 7.36MNPFIRR7 pKa = 11.84LGSLLMGKK15 pKa = 6.16TTKK18 pKa = 10.47EE19 pKa = 4.05LIIGLYY25 pKa = 10.26SCVQTLHH32 pKa = 7.06KK33 pKa = 10.06IYY35 pKa = 10.37LSQGSRR41 pKa = 11.84GLAIRR46 pKa = 11.84LKK48 pKa = 10.79ASSTYY53 pKa = 11.03LMKK56 pKa = 10.5SWSGDD61 pKa = 3.37PLLDD65 pKa = 3.93CKK67 pKa = 11.2AFGPTMSLTPRR78 pKa = 11.84GIPIWIPSAWRR89 pKa = 11.84CQISSRR95 pKa = 11.84NLAIFRR101 pKa = 11.84VTMTLCNLYY110 pKa = 10.22RR111 pKa = 11.84VLPYY115 pKa = 10.07QGKK118 pKa = 9.84LKK120 pKa = 10.8LSTITDD126 pKa = 3.59DD127 pKa = 4.69RR128 pKa = 11.84KK129 pKa = 11.09GSTPDD134 pKa = 3.68LMFSFIKK141 pKa = 10.71DD142 pKa = 3.1KK143 pKa = 10.62FAPNITNRR151 pKa = 11.84VPYY154 pKa = 9.86PGIFSWDD161 pKa = 3.66PKK163 pKa = 10.32PLLTRR168 pKa = 11.84GPGAQKK174 pKa = 10.5EE175 pKa = 4.37KK176 pKa = 10.08NTMSGFGMALRR187 pKa = 11.84AIHH190 pKa = 5.34EE191 pKa = 4.43TGMLDD196 pKa = 3.51HH197 pKa = 6.92LKK199 pKa = 10.57AYY201 pKa = 10.49ALYY204 pKa = 10.01TGLNPQLEE212 pKa = 4.49SLKK215 pKa = 10.78VIWEE219 pKa = 4.43GTRR222 pKa = 11.84KK223 pKa = 9.49HH224 pKa = 6.48CSIGPIGKK232 pKa = 9.04LAFKK236 pKa = 10.41QEE238 pKa = 4.09PGKK241 pKa = 10.85VRR243 pKa = 11.84VFAMVDD249 pKa = 4.5CITQWFLHH257 pKa = 6.46PLHH260 pKa = 7.0KK261 pKa = 10.84YY262 pKa = 9.69LFSVLRR268 pKa = 11.84TTKK271 pKa = 10.5EE272 pKa = 3.65DD273 pKa = 3.2ATFDD277 pKa = 3.55QEE279 pKa = 4.6KK280 pKa = 10.45GINLVRR286 pKa = 11.84LALSKK291 pKa = 11.14KK292 pKa = 9.26LDD294 pKa = 3.41KK295 pKa = 11.2SVFSFDD301 pKa = 4.34LSAATDD307 pKa = 4.13RR308 pKa = 11.84LPMDD312 pKa = 3.3IQMVILNSLTPFWLAEE328 pKa = 3.75KK329 pKa = 10.26AVRR332 pKa = 11.84HH333 pKa = 5.04GVKK336 pKa = 10.43GGLGDD341 pKa = 3.64AWADD345 pKa = 3.57LLVDD349 pKa = 3.81RR350 pKa = 11.84DD351 pKa = 4.02YY352 pKa = 11.8YY353 pKa = 10.39LPRR356 pKa = 11.84WSGYY360 pKa = 10.65ARR362 pKa = 11.84DD363 pKa = 3.79SKK365 pKa = 11.13VRR367 pKa = 11.84YY368 pKa = 9.03AVGQPMGALSSWAMLALTHH387 pKa = 6.72HH388 pKa = 6.94MIVQFAAASVGVTGWFKK405 pKa = 11.06EE406 pKa = 3.79YY407 pKa = 9.9MVLGDD412 pKa = 5.78DD413 pKa = 3.29IVIYY417 pKa = 10.56NSEE420 pKa = 3.96VAKK423 pKa = 10.58AYY425 pKa = 7.24STLMGTLGVGISDD438 pKa = 3.93TKK440 pKa = 11.32SLTSKK445 pKa = 10.23IGVFEE450 pKa = 3.99FAKK453 pKa = 10.55RR454 pKa = 11.84LMDD457 pKa = 5.63LEE459 pKa = 5.37GPCQGLPLAEE469 pKa = 4.04FAAARR474 pKa = 11.84FNLSILFQSFRR485 pKa = 11.84SRR487 pKa = 11.84TLYY490 pKa = 10.25PKK492 pKa = 9.83ISTFMRR498 pKa = 11.84FLGFGYY504 pKa = 10.21KK505 pKa = 10.08VLGSLGMRR513 pKa = 11.84LGDD516 pKa = 3.25MRR518 pKa = 11.84KK519 pKa = 9.03RR520 pKa = 11.84SGYY523 pKa = 10.03FEE525 pKa = 4.19TVAYY529 pKa = 10.09SPLVTDD535 pKa = 4.13KK536 pKa = 11.25SVNKK540 pKa = 9.15WSEE543 pKa = 3.63FFKK546 pKa = 10.95RR547 pKa = 11.84FSTYY551 pKa = 10.99DD552 pKa = 3.68LYY554 pKa = 10.7PLVEE558 pKa = 4.11ACVYY562 pKa = 9.98KK563 pKa = 10.48AYY565 pKa = 10.71HH566 pKa = 7.23LIPALTKK573 pKa = 9.9FDD575 pKa = 3.9RR576 pKa = 11.84WNLWRR581 pKa = 11.84TLFPHH586 pKa = 7.01FDD588 pKa = 3.63DD589 pKa = 5.68KK590 pKa = 11.43KK591 pKa = 10.08ILLIPEE597 pKa = 3.75VAGRR601 pKa = 11.84HH602 pKa = 5.26LEE604 pKa = 4.12SLFTSMLDD612 pKa = 3.49NKK614 pKa = 8.67VAKK617 pKa = 10.24YY618 pKa = 10.22KK619 pKa = 10.4ADD621 pKa = 2.98HH622 pKa = 5.86SVYY625 pKa = 10.69VNRR628 pKa = 11.84LNRR631 pKa = 11.84AEE633 pKa = 4.32VPTNDD638 pKa = 3.2WEE640 pKa = 5.92AIDD643 pKa = 3.85TLLSIMEE650 pKa = 4.42TSPPTSDD657 pKa = 5.05LLQDD661 pKa = 3.59PDD663 pKa = 3.8MNEE666 pKa = 3.88LGFGLSVSNYY676 pKa = 10.82DD677 pKa = 3.89EE678 pKa = 4.99DD679 pKa = 4.6NKK681 pKa = 11.28SKK683 pKa = 9.13TCSLSLPYY691 pKa = 10.13TVFTKK696 pKa = 10.29IPQLLDD702 pKa = 4.64SIDD705 pKa = 3.9DD706 pKa = 3.6WVEE709 pKa = 3.61PIIPDD714 pKa = 3.25KK715 pKa = 11.01HH716 pKa = 6.99PEE718 pKa = 3.59VSADD722 pKa = 3.33VNRR725 pKa = 11.84PLIHH729 pKa = 6.83KK730 pKa = 8.61PGKK733 pKa = 10.42VFVKK737 pKa = 10.46GIATGKK743 pKa = 9.91NEE745 pKa = 3.99NNKK748 pKa = 9.43VEE750 pKa = 4.23ITKK753 pKa = 9.28MLQDD757 pKa = 4.09KK758 pKa = 10.71KK759 pKa = 10.17FLAYY763 pKa = 9.5IIEE766 pKa = 4.4SGKK769 pKa = 8.25ITIYY773 pKa = 10.87SPAFQAMIRR782 pKa = 11.84ASWAKK787 pKa = 9.89AQSGSKK793 pKa = 10.14
Molecular weight: 89.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
793 |
793 |
793 |
793.0 |
89.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.683 ± 0.0 | 1.135 ± 0.0 |
5.675 ± 0.0 | 3.657 ± 0.0 |
5.044 ± 0.0 | 6.557 ± 0.0 |
1.892 ± 0.0 | 5.801 ± 0.0 |
7.818 ± 0.0 | 11.475 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.405 ± 0.0 | 3.026 ± 0.0 |
5.296 ± 0.0 | 2.396 ± 0.0 |
4.792 ± 0.0 | 7.945 ± 0.0 |
5.927 ± 0.0 | 5.549 ± 0.0 |
2.144 ± 0.0 | 3.783 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
