
Hubei tombus-like virus 28
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
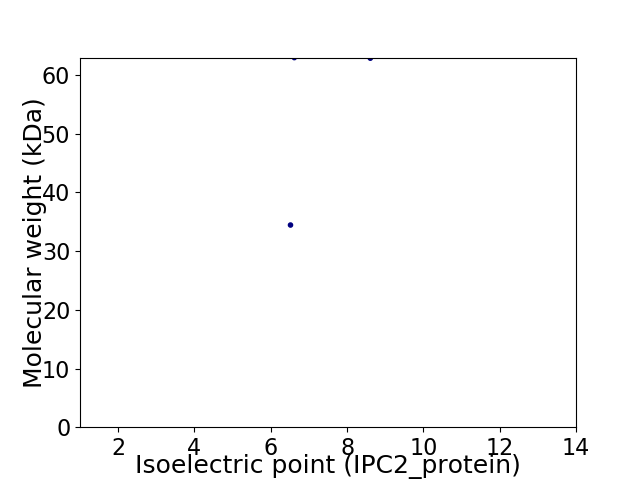
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
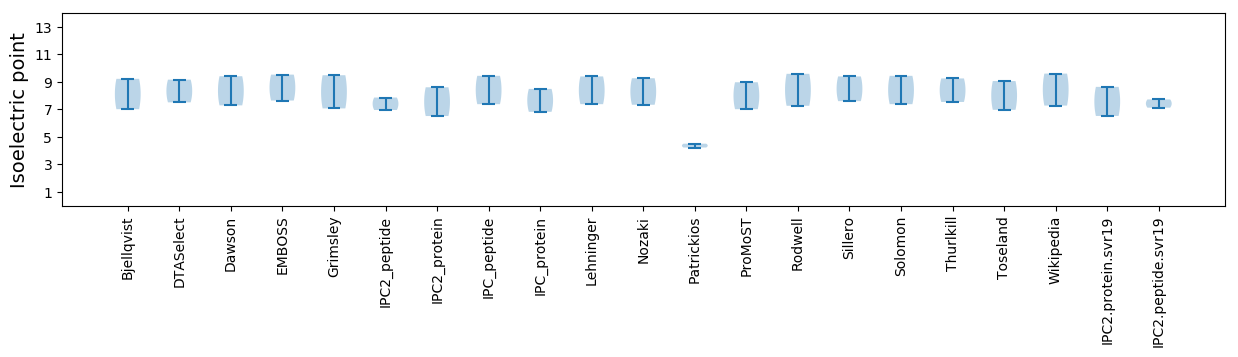
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGB3|A0A1L3KGB3_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 28 OX=1923275 PE=4 SV=1
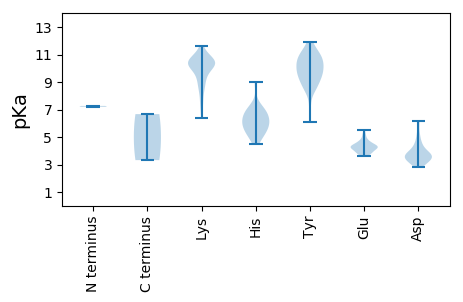
MM1 pKa = 7.26SVYY4 pKa = 9.84TGYY7 pKa = 8.59YY8 pKa = 8.57TPNNTTWYY16 pKa = 9.68QRR18 pKa = 11.84IGEE21 pKa = 4.31WVRR24 pKa = 11.84DD25 pKa = 3.46AMNYY29 pKa = 9.44NPEE32 pKa = 4.43LKK34 pKa = 10.15RR35 pKa = 11.84HH36 pKa = 5.09EE37 pKa = 4.84DD38 pKa = 3.19KK39 pKa = 11.59VEE41 pKa = 3.8EE42 pKa = 4.29LKK44 pKa = 11.31LKK46 pKa = 9.0ITEE49 pKa = 4.1NQNRR53 pKa = 11.84LLEE56 pKa = 4.65AEE58 pKa = 4.65LDD60 pKa = 3.6QHH62 pKa = 7.19AKK64 pKa = 10.55ALEE67 pKa = 4.17LDD69 pKa = 3.69AMKK72 pKa = 10.49VDD74 pKa = 3.73HH75 pKa = 7.29KK76 pKa = 11.07EE77 pKa = 3.64GLLAHH82 pKa = 6.75LKK84 pKa = 10.55FYY86 pKa = 10.28TLNKK90 pKa = 10.24KK91 pKa = 9.66IDD93 pKa = 3.76TKK95 pKa = 11.24YY96 pKa = 9.76IDD98 pKa = 3.64YY99 pKa = 8.62VTRR102 pKa = 11.84LAEE105 pKa = 4.45AYY107 pKa = 9.93FEE109 pKa = 4.32SMKK112 pKa = 10.48IKK114 pKa = 10.52NPRR117 pKa = 11.84LKK119 pKa = 10.62CEE121 pKa = 4.27LMDD124 pKa = 3.86EE125 pKa = 4.31VLPIHH130 pKa = 6.74IKK132 pKa = 10.17QRR134 pKa = 11.84LSTMPLLMKK143 pKa = 10.3DD144 pKa = 3.57DD145 pKa = 3.86RR146 pKa = 11.84TIASANYY153 pKa = 8.9ISEE156 pKa = 5.13LIDD159 pKa = 3.47GRR161 pKa = 11.84KK162 pKa = 9.34VSYY165 pKa = 9.34PWWNLWQPTYY175 pKa = 11.36ARR177 pKa = 11.84LGGSGNEE184 pKa = 3.98NADD187 pKa = 3.62YY188 pKa = 10.84LVPTQPNFWKK198 pKa = 10.64YY199 pKa = 11.0GMIVTGSVAIALGGKK214 pKa = 8.8WIISRR219 pKa = 11.84ASQNLTDD226 pKa = 6.04GITNLLAQKK235 pKa = 10.01AQQLPSTTVITQSLSKK251 pKa = 11.11DD252 pKa = 3.46MIEE255 pKa = 4.33IASKK259 pKa = 10.48GCTNIYY265 pKa = 10.86EE266 pKa = 4.28GMLWIWNSGHH276 pKa = 6.19VSNILPAFGQQASDD290 pKa = 3.18EE291 pKa = 4.63SIRR294 pKa = 11.84MIMTSFF300 pKa = 3.32
MM1 pKa = 7.26SVYY4 pKa = 9.84TGYY7 pKa = 8.59YY8 pKa = 8.57TPNNTTWYY16 pKa = 9.68QRR18 pKa = 11.84IGEE21 pKa = 4.31WVRR24 pKa = 11.84DD25 pKa = 3.46AMNYY29 pKa = 9.44NPEE32 pKa = 4.43LKK34 pKa = 10.15RR35 pKa = 11.84HH36 pKa = 5.09EE37 pKa = 4.84DD38 pKa = 3.19KK39 pKa = 11.59VEE41 pKa = 3.8EE42 pKa = 4.29LKK44 pKa = 11.31LKK46 pKa = 9.0ITEE49 pKa = 4.1NQNRR53 pKa = 11.84LLEE56 pKa = 4.65AEE58 pKa = 4.65LDD60 pKa = 3.6QHH62 pKa = 7.19AKK64 pKa = 10.55ALEE67 pKa = 4.17LDD69 pKa = 3.69AMKK72 pKa = 10.49VDD74 pKa = 3.73HH75 pKa = 7.29KK76 pKa = 11.07EE77 pKa = 3.64GLLAHH82 pKa = 6.75LKK84 pKa = 10.55FYY86 pKa = 10.28TLNKK90 pKa = 10.24KK91 pKa = 9.66IDD93 pKa = 3.76TKK95 pKa = 11.24YY96 pKa = 9.76IDD98 pKa = 3.64YY99 pKa = 8.62VTRR102 pKa = 11.84LAEE105 pKa = 4.45AYY107 pKa = 9.93FEE109 pKa = 4.32SMKK112 pKa = 10.48IKK114 pKa = 10.52NPRR117 pKa = 11.84LKK119 pKa = 10.62CEE121 pKa = 4.27LMDD124 pKa = 3.86EE125 pKa = 4.31VLPIHH130 pKa = 6.74IKK132 pKa = 10.17QRR134 pKa = 11.84LSTMPLLMKK143 pKa = 10.3DD144 pKa = 3.57DD145 pKa = 3.86RR146 pKa = 11.84TIASANYY153 pKa = 8.9ISEE156 pKa = 5.13LIDD159 pKa = 3.47GRR161 pKa = 11.84KK162 pKa = 9.34VSYY165 pKa = 9.34PWWNLWQPTYY175 pKa = 11.36ARR177 pKa = 11.84LGGSGNEE184 pKa = 3.98NADD187 pKa = 3.62YY188 pKa = 10.84LVPTQPNFWKK198 pKa = 10.64YY199 pKa = 11.0GMIVTGSVAIALGGKK214 pKa = 8.8WIISRR219 pKa = 11.84ASQNLTDD226 pKa = 6.04GITNLLAQKK235 pKa = 10.01AQQLPSTTVITQSLSKK251 pKa = 11.11DD252 pKa = 3.46MIEE255 pKa = 4.33IASKK259 pKa = 10.48GCTNIYY265 pKa = 10.86EE266 pKa = 4.28GMLWIWNSGHH276 pKa = 6.19VSNILPAFGQQASDD290 pKa = 3.18EE291 pKa = 4.63SIRR294 pKa = 11.84MIMTSFF300 pKa = 3.32
Molecular weight: 34.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGB3|A0A1L3KGB3_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 28 OX=1923275 PE=4 SV=1
MM1 pKa = 7.2VEE3 pKa = 5.24LMATDD8 pKa = 4.4LCQTGWIGKK17 pKa = 8.31RR18 pKa = 11.84EE19 pKa = 4.07CGLPRR24 pKa = 11.84THH26 pKa = 6.02TTQFLEE32 pKa = 3.97IRR34 pKa = 11.84NDD36 pKa = 3.07SYY38 pKa = 11.66RR39 pKa = 11.84IGSHH43 pKa = 5.94RR44 pKa = 11.84FGRR47 pKa = 11.84QMDD50 pKa = 4.22YY51 pKa = 10.59FPSIAKK57 pKa = 9.6PDD59 pKa = 2.95GWYY62 pKa = 10.26YY63 pKa = 10.94EE64 pKa = 4.56PIGSEE69 pKa = 3.81SAAAAFYY76 pKa = 10.66NRR78 pKa = 11.84HH79 pKa = 5.32NPEE82 pKa = 4.76LIEE85 pKa = 5.05GYY87 pKa = 10.2DD88 pKa = 3.56RR89 pKa = 11.84NCIKK93 pKa = 10.7GLYY96 pKa = 8.33KK97 pKa = 10.15HH98 pKa = 6.11LRR100 pKa = 11.84RR101 pKa = 11.84YY102 pKa = 8.69VVDD105 pKa = 3.66MEE107 pKa = 4.1QWTCEE112 pKa = 3.85QYY114 pKa = 10.54IASIWPASKK123 pKa = 10.39RR124 pKa = 11.84RR125 pKa = 11.84IYY127 pKa = 10.49QDD129 pKa = 5.34DD130 pKa = 3.83YY131 pKa = 11.95DD132 pKa = 4.12QFLSDD137 pKa = 4.25GKK139 pKa = 10.45ISSLVQPFTKK149 pKa = 10.03PEE151 pKa = 3.76KK152 pKa = 10.67KK153 pKa = 10.38NFSKK157 pKa = 11.09YY158 pKa = 9.4KK159 pKa = 8.81APRR162 pKa = 11.84MIQARR167 pKa = 11.84KK168 pKa = 8.99PIFNIHH174 pKa = 5.52YY175 pKa = 9.87GKK177 pKa = 10.4FIKK180 pKa = 10.23PLEE183 pKa = 4.08QKK185 pKa = 8.31ITKK188 pKa = 9.48HH189 pKa = 5.22GRR191 pKa = 11.84FRR193 pKa = 11.84DD194 pKa = 3.78HH195 pKa = 6.97FGKK198 pKa = 10.84GSPDD202 pKa = 3.18EE203 pKa = 3.79IARR206 pKa = 11.84RR207 pKa = 11.84ITKK210 pKa = 10.58LSLKK214 pKa = 8.63WQYY217 pKa = 9.02KK218 pKa = 8.29TEE220 pKa = 4.31GDD222 pKa = 3.49HH223 pKa = 5.52TTFDD227 pKa = 3.16AHH229 pKa = 5.58ITKK232 pKa = 10.22EE233 pKa = 4.08LLEE236 pKa = 4.43LTHH239 pKa = 6.8KK240 pKa = 10.68FYY242 pKa = 10.96AACYY246 pKa = 9.29NNNPEE251 pKa = 4.78LKK253 pKa = 9.46RR254 pKa = 11.84LSKK257 pKa = 9.82RR258 pKa = 11.84TIINRR263 pKa = 11.84CRR265 pKa = 11.84SRR267 pKa = 11.84HH268 pKa = 4.48GDD270 pKa = 2.85KK271 pKa = 10.19YY272 pKa = 8.56TVVGSRR278 pKa = 11.84MSGDD282 pKa = 3.12VDD284 pKa = 3.53TSFGNSIINIAILSEE299 pKa = 4.11LLYY302 pKa = 11.07KK303 pKa = 10.82LGINGEE309 pKa = 4.41FIVNGDD315 pKa = 3.78DD316 pKa = 6.19FILFTNKK323 pKa = 9.36PVNIKK328 pKa = 8.92MAEE331 pKa = 4.26AILRR335 pKa = 11.84TMNMEE340 pKa = 4.32TKK342 pKa = 9.61MKK344 pKa = 10.27EE345 pKa = 3.81STEE348 pKa = 4.22NIHH351 pKa = 6.2TVEE354 pKa = 4.64FCQTKK359 pKa = 10.36LVLTAEE365 pKa = 4.3GRR367 pKa = 11.84YY368 pKa = 6.09TTMKK372 pKa = 10.19HH373 pKa = 4.88IRR375 pKa = 11.84KK376 pKa = 7.08TVEE379 pKa = 3.63KK380 pKa = 10.42FGMTFVTTRR389 pKa = 11.84NYY391 pKa = 9.53WIYY394 pKa = 11.0VLEE397 pKa = 4.53NAHH400 pKa = 6.1GMWMMNKK407 pKa = 7.28TTPLGLLFKK416 pKa = 10.5HH417 pKa = 6.79IYY419 pKa = 10.26FKK421 pKa = 10.51MIEE424 pKa = 4.36LDD426 pKa = 3.37MVPNAKK432 pKa = 9.94EE433 pKa = 4.41LINKK437 pKa = 7.54HH438 pKa = 5.7HH439 pKa = 6.56RR440 pKa = 11.84LEE442 pKa = 4.09YY443 pKa = 10.39EE444 pKa = 3.83YY445 pKa = 11.25LEE447 pKa = 4.04RR448 pKa = 11.84AFIYY452 pKa = 9.5FIKK455 pKa = 10.54LARR458 pKa = 11.84KK459 pKa = 7.76DD460 pKa = 3.34TAMSDD465 pKa = 3.4DD466 pKa = 3.81HH467 pKa = 9.02SEE469 pKa = 3.84ITISMIAADD478 pKa = 4.76EE479 pKa = 4.33DD480 pKa = 4.47VMSLDD485 pKa = 4.66EE486 pKa = 5.51IEE488 pKa = 4.18QQLMQRR494 pKa = 11.84IIKK497 pKa = 10.03LYY499 pKa = 10.17NRR501 pKa = 11.84YY502 pKa = 7.19PTLSTKK508 pKa = 10.19QLLSMPYY515 pKa = 10.17NRR517 pKa = 11.84NLYY520 pKa = 9.43IDD522 pKa = 5.13HH523 pKa = 6.25NTKK526 pKa = 6.38TTPYY530 pKa = 9.48FVNHH534 pKa = 6.67
MM1 pKa = 7.2VEE3 pKa = 5.24LMATDD8 pKa = 4.4LCQTGWIGKK17 pKa = 8.31RR18 pKa = 11.84EE19 pKa = 4.07CGLPRR24 pKa = 11.84THH26 pKa = 6.02TTQFLEE32 pKa = 3.97IRR34 pKa = 11.84NDD36 pKa = 3.07SYY38 pKa = 11.66RR39 pKa = 11.84IGSHH43 pKa = 5.94RR44 pKa = 11.84FGRR47 pKa = 11.84QMDD50 pKa = 4.22YY51 pKa = 10.59FPSIAKK57 pKa = 9.6PDD59 pKa = 2.95GWYY62 pKa = 10.26YY63 pKa = 10.94EE64 pKa = 4.56PIGSEE69 pKa = 3.81SAAAAFYY76 pKa = 10.66NRR78 pKa = 11.84HH79 pKa = 5.32NPEE82 pKa = 4.76LIEE85 pKa = 5.05GYY87 pKa = 10.2DD88 pKa = 3.56RR89 pKa = 11.84NCIKK93 pKa = 10.7GLYY96 pKa = 8.33KK97 pKa = 10.15HH98 pKa = 6.11LRR100 pKa = 11.84RR101 pKa = 11.84YY102 pKa = 8.69VVDD105 pKa = 3.66MEE107 pKa = 4.1QWTCEE112 pKa = 3.85QYY114 pKa = 10.54IASIWPASKK123 pKa = 10.39RR124 pKa = 11.84RR125 pKa = 11.84IYY127 pKa = 10.49QDD129 pKa = 5.34DD130 pKa = 3.83YY131 pKa = 11.95DD132 pKa = 4.12QFLSDD137 pKa = 4.25GKK139 pKa = 10.45ISSLVQPFTKK149 pKa = 10.03PEE151 pKa = 3.76KK152 pKa = 10.67KK153 pKa = 10.38NFSKK157 pKa = 11.09YY158 pKa = 9.4KK159 pKa = 8.81APRR162 pKa = 11.84MIQARR167 pKa = 11.84KK168 pKa = 8.99PIFNIHH174 pKa = 5.52YY175 pKa = 9.87GKK177 pKa = 10.4FIKK180 pKa = 10.23PLEE183 pKa = 4.08QKK185 pKa = 8.31ITKK188 pKa = 9.48HH189 pKa = 5.22GRR191 pKa = 11.84FRR193 pKa = 11.84DD194 pKa = 3.78HH195 pKa = 6.97FGKK198 pKa = 10.84GSPDD202 pKa = 3.18EE203 pKa = 3.79IARR206 pKa = 11.84RR207 pKa = 11.84ITKK210 pKa = 10.58LSLKK214 pKa = 8.63WQYY217 pKa = 9.02KK218 pKa = 8.29TEE220 pKa = 4.31GDD222 pKa = 3.49HH223 pKa = 5.52TTFDD227 pKa = 3.16AHH229 pKa = 5.58ITKK232 pKa = 10.22EE233 pKa = 4.08LLEE236 pKa = 4.43LTHH239 pKa = 6.8KK240 pKa = 10.68FYY242 pKa = 10.96AACYY246 pKa = 9.29NNNPEE251 pKa = 4.78LKK253 pKa = 9.46RR254 pKa = 11.84LSKK257 pKa = 9.82RR258 pKa = 11.84TIINRR263 pKa = 11.84CRR265 pKa = 11.84SRR267 pKa = 11.84HH268 pKa = 4.48GDD270 pKa = 2.85KK271 pKa = 10.19YY272 pKa = 8.56TVVGSRR278 pKa = 11.84MSGDD282 pKa = 3.12VDD284 pKa = 3.53TSFGNSIINIAILSEE299 pKa = 4.11LLYY302 pKa = 11.07KK303 pKa = 10.82LGINGEE309 pKa = 4.41FIVNGDD315 pKa = 3.78DD316 pKa = 6.19FILFTNKK323 pKa = 9.36PVNIKK328 pKa = 8.92MAEE331 pKa = 4.26AILRR335 pKa = 11.84TMNMEE340 pKa = 4.32TKK342 pKa = 9.61MKK344 pKa = 10.27EE345 pKa = 3.81STEE348 pKa = 4.22NIHH351 pKa = 6.2TVEE354 pKa = 4.64FCQTKK359 pKa = 10.36LVLTAEE365 pKa = 4.3GRR367 pKa = 11.84YY368 pKa = 6.09TTMKK372 pKa = 10.19HH373 pKa = 4.88IRR375 pKa = 11.84KK376 pKa = 7.08TVEE379 pKa = 3.63KK380 pKa = 10.42FGMTFVTTRR389 pKa = 11.84NYY391 pKa = 9.53WIYY394 pKa = 11.0VLEE397 pKa = 4.53NAHH400 pKa = 6.1GMWMMNKK407 pKa = 7.28TTPLGLLFKK416 pKa = 10.5HH417 pKa = 6.79IYY419 pKa = 10.26FKK421 pKa = 10.51MIEE424 pKa = 4.36LDD426 pKa = 3.37MVPNAKK432 pKa = 9.94EE433 pKa = 4.41LINKK437 pKa = 7.54HH438 pKa = 5.7HH439 pKa = 6.56RR440 pKa = 11.84LEE442 pKa = 4.09YY443 pKa = 10.39EE444 pKa = 3.83YY445 pKa = 11.25LEE447 pKa = 4.04RR448 pKa = 11.84AFIYY452 pKa = 9.5FIKK455 pKa = 10.54LARR458 pKa = 11.84KK459 pKa = 7.76DD460 pKa = 3.34TAMSDD465 pKa = 3.4DD466 pKa = 3.81HH467 pKa = 9.02SEE469 pKa = 3.84ITISMIAADD478 pKa = 4.76EE479 pKa = 4.33DD480 pKa = 4.47VMSLDD485 pKa = 4.66EE486 pKa = 5.51IEE488 pKa = 4.18QQLMQRR494 pKa = 11.84IIKK497 pKa = 10.03LYY499 pKa = 10.17NRR501 pKa = 11.84YY502 pKa = 7.19PTLSTKK508 pKa = 10.19QLLSMPYY515 pKa = 10.17NRR517 pKa = 11.84NLYY520 pKa = 9.43IDD522 pKa = 5.13HH523 pKa = 6.25NTKK526 pKa = 6.38TTPYY530 pKa = 9.48FVNHH534 pKa = 6.67
Molecular weight: 62.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
834 |
300 |
534 |
417.0 |
48.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.396 ± 0.739 | 1.079 ± 0.24 |
5.036 ± 0.021 | 6.475 ± 0.082 |
3.597 ± 1.122 | 5.036 ± 0.173 |
3.118 ± 0.65 | 8.153 ± 0.283 |
7.914 ± 0.337 | 8.633 ± 0.988 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.077 ± 0.045 | 5.516 ± 0.282 |
3.597 ± 0.04 | 3.477 ± 0.498 |
5.516 ± 0.881 | 5.516 ± 0.475 |
7.074 ± 0.237 | 3.477 ± 0.304 |
1.918 ± 0.629 | 5.396 ± 0.23 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
