
Sandaracinobacter sp. PAMC 28131
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingosinicellaceae; Sandaracinobacter; unclassified Sandaracinobacter
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
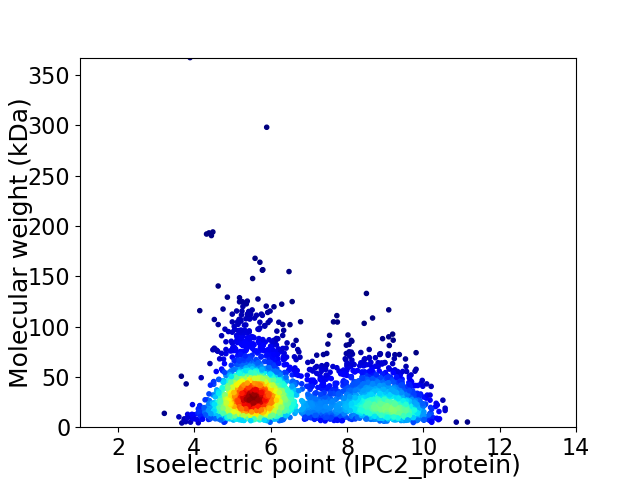
Virtual 2D-PAGE plot for 3327 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
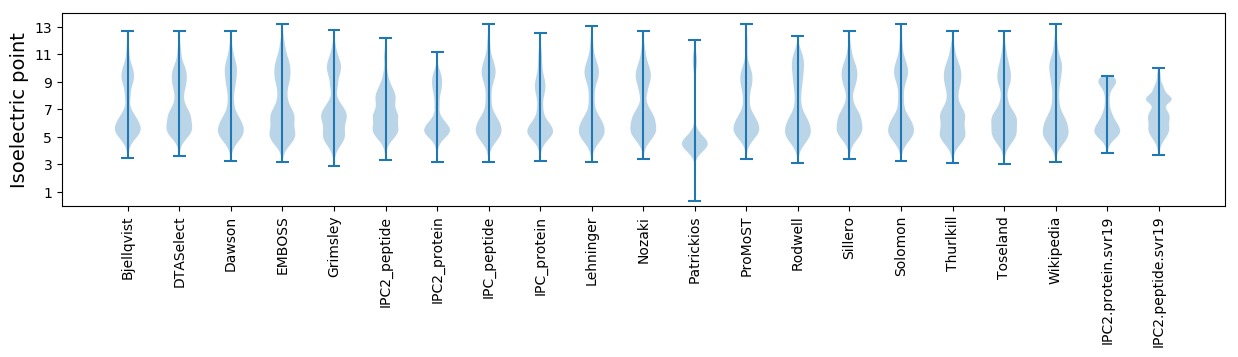
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A501XNG0|A0A501XNG0_9SPHN Cysteine hydrolase OS=Sandaracinobacter sp. PAMC 28131 OX=1715348 GN=FJQ54_06625 PE=4 SV=1
MM1 pKa = 8.06RR2 pKa = 11.84ILTGLLLGCASFVALSSTPASAATCFGNCGEE33 pKa = 4.3TNINGGQTGSVGVPPLQGDD52 pKa = 3.64NVYY55 pKa = 10.95YY56 pKa = 9.94WVSTNGGSTGGGTLPGQEE74 pKa = 4.19SGSTNGSRR82 pKa = 11.84LVSDD86 pKa = 4.07SFYY89 pKa = 11.18AAAGQKK95 pKa = 9.47VSYY98 pKa = 8.57YY99 pKa = 10.46FNYY102 pKa = 8.15ITSDD106 pKa = 3.14GAGYY110 pKa = 11.0ADD112 pKa = 4.27YY113 pKa = 10.89SWSQLRR119 pKa = 11.84DD120 pKa = 3.46VNDD123 pKa = 3.54SSNVTNLFTARR134 pKa = 11.84TKK136 pKa = 10.63PSGTIVPGLDD146 pKa = 3.55LPDD149 pKa = 3.81VEE151 pKa = 5.15ATLSPATVLIKK162 pKa = 10.6PGTTFAPLGDD172 pKa = 3.68SSGACWSAGCGNTGWIYY189 pKa = 11.07SEE191 pKa = 4.12YY192 pKa = 10.04TIQTSGTYY200 pKa = 9.65EE201 pKa = 3.95LVFGVSNWDD210 pKa = 3.16DD211 pKa = 3.56TAFDD215 pKa = 3.54SALAFDD221 pKa = 4.62GLLIGGIVIGDD232 pKa = 3.97GSSPTNPLAPTDD244 pKa = 4.12LQPDD248 pKa = 4.12GSFVFSFEE256 pKa = 4.05VTQPEE261 pKa = 4.2QPVFIDD267 pKa = 3.98PYY269 pKa = 8.7VAVGYY274 pKa = 9.49EE275 pKa = 3.92YY276 pKa = 10.9EE277 pKa = 4.44VIDD280 pKa = 4.19GSLAFSQAWFGDD292 pKa = 3.31VGDD295 pKa = 4.37PDD297 pKa = 5.65GYY299 pKa = 10.75QIWGWDD305 pKa = 3.2GADD308 pKa = 3.49YY309 pKa = 11.47YY310 pKa = 11.43FLTDD314 pKa = 3.32ITAEE318 pKa = 3.65TWYY321 pKa = 10.83AFASPVTKK329 pKa = 10.65FKK331 pKa = 11.24LLGIDD336 pKa = 3.69SDD338 pKa = 4.59LGLDD342 pKa = 3.95PEE344 pKa = 4.98DD345 pKa = 4.14PNAFVSGFIYY355 pKa = 10.73NGTGKK360 pKa = 10.1VDD362 pKa = 3.63VSMKK366 pKa = 10.5PITEE370 pKa = 4.02WVEE373 pKa = 3.92GGGGGVVPEE382 pKa = 4.08PATWAMMILGFGAVGLAARR401 pKa = 11.84RR402 pKa = 11.84RR403 pKa = 11.84RR404 pKa = 11.84SHH406 pKa = 6.59SVTAA410 pKa = 4.29
MM1 pKa = 8.06RR2 pKa = 11.84ILTGLLLGCASFVALSSTPASAATCFGNCGEE33 pKa = 4.3TNINGGQTGSVGVPPLQGDD52 pKa = 3.64NVYY55 pKa = 10.95YY56 pKa = 9.94WVSTNGGSTGGGTLPGQEE74 pKa = 4.19SGSTNGSRR82 pKa = 11.84LVSDD86 pKa = 4.07SFYY89 pKa = 11.18AAAGQKK95 pKa = 9.47VSYY98 pKa = 8.57YY99 pKa = 10.46FNYY102 pKa = 8.15ITSDD106 pKa = 3.14GAGYY110 pKa = 11.0ADD112 pKa = 4.27YY113 pKa = 10.89SWSQLRR119 pKa = 11.84DD120 pKa = 3.46VNDD123 pKa = 3.54SSNVTNLFTARR134 pKa = 11.84TKK136 pKa = 10.63PSGTIVPGLDD146 pKa = 3.55LPDD149 pKa = 3.81VEE151 pKa = 5.15ATLSPATVLIKK162 pKa = 10.6PGTTFAPLGDD172 pKa = 3.68SSGACWSAGCGNTGWIYY189 pKa = 11.07SEE191 pKa = 4.12YY192 pKa = 10.04TIQTSGTYY200 pKa = 9.65EE201 pKa = 3.95LVFGVSNWDD210 pKa = 3.16DD211 pKa = 3.56TAFDD215 pKa = 3.54SALAFDD221 pKa = 4.62GLLIGGIVIGDD232 pKa = 3.97GSSPTNPLAPTDD244 pKa = 4.12LQPDD248 pKa = 4.12GSFVFSFEE256 pKa = 4.05VTQPEE261 pKa = 4.2QPVFIDD267 pKa = 3.98PYY269 pKa = 8.7VAVGYY274 pKa = 9.49EE275 pKa = 3.92YY276 pKa = 10.9EE277 pKa = 4.44VIDD280 pKa = 4.19GSLAFSQAWFGDD292 pKa = 3.31VGDD295 pKa = 4.37PDD297 pKa = 5.65GYY299 pKa = 10.75QIWGWDD305 pKa = 3.2GADD308 pKa = 3.49YY309 pKa = 11.47YY310 pKa = 11.43FLTDD314 pKa = 3.32ITAEE318 pKa = 3.65TWYY321 pKa = 10.83AFASPVTKK329 pKa = 10.65FKK331 pKa = 11.24LLGIDD336 pKa = 3.69SDD338 pKa = 4.59LGLDD342 pKa = 3.95PEE344 pKa = 4.98DD345 pKa = 4.14PNAFVSGFIYY355 pKa = 10.73NGTGKK360 pKa = 10.1VDD362 pKa = 3.63VSMKK366 pKa = 10.5PITEE370 pKa = 4.02WVEE373 pKa = 3.92GGGGGVVPEE382 pKa = 4.08PATWAMMILGFGAVGLAARR401 pKa = 11.84RR402 pKa = 11.84RR403 pKa = 11.84RR404 pKa = 11.84SHH406 pKa = 6.59SVTAA410 pKa = 4.29
Molecular weight: 43.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A501XDP2|A0A501XDP2_9SPHN 3'(2') 5'-bisphosphate nucleotidase CysQ OS=Sandaracinobacter sp. PAMC 28131 OX=1715348 GN=cysQ PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.39GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTVGGRR28 pKa = 11.84KK29 pKa = 8.56VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.76VLSAA44 pKa = 4.11
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.39GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTVGGRR28 pKa = 11.84KK29 pKa = 8.56VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.76VLSAA44 pKa = 4.11
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1063820 |
37 |
3595 |
319.8 |
34.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.694 ± 0.061 | 0.674 ± 0.011 |
5.574 ± 0.029 | 5.369 ± 0.045 |
3.505 ± 0.025 | 9.058 ± 0.083 |
1.938 ± 0.02 | 4.729 ± 0.027 |
2.939 ± 0.033 | 10.529 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.328 ± 0.022 | 2.477 ± 0.031 |
5.698 ± 0.04 | 3.218 ± 0.026 |
7.299 ± 0.053 | 5.335 ± 0.038 |
5.142 ± 0.039 | 7.013 ± 0.039 |
1.484 ± 0.019 | 1.999 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
