
Bos taurus papillomavirus 15
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Xipapillomavirus; Xipapillomavirus 1
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
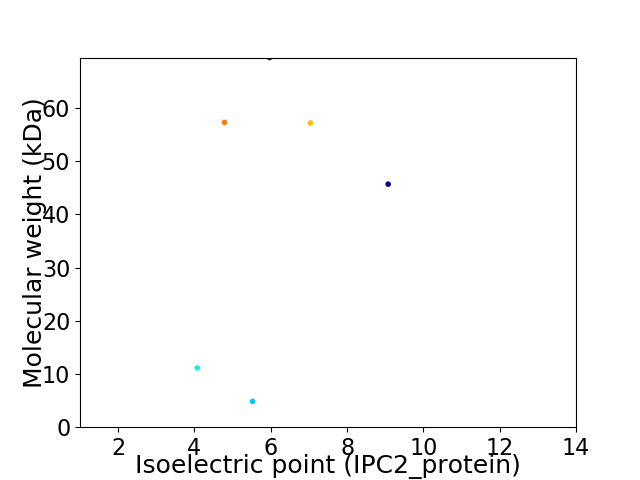
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5JF74|A0A0B5JF74_BPV3 E8 protein OS=Bos taurus papillomavirus 15 OX=1969700 GN=E5 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 10.38GQDD5 pKa = 3.51VVLQDD10 pKa = 4.08LASEE14 pKa = 4.65LEE16 pKa = 4.95DD17 pKa = 3.08IVSPINLDD25 pKa = 3.47CEE27 pKa = 4.39EE28 pKa = 4.55EE29 pKa = 4.24IEE31 pKa = 4.29TEE33 pKa = 4.04EE34 pKa = 5.12VEE36 pKa = 4.47VDD38 pKa = 4.11FPNPYY43 pKa = 10.16EE44 pKa = 4.09ITAVCYY50 pKa = 10.43VCEE53 pKa = 4.03EE54 pKa = 4.21TLRR57 pKa = 11.84IAVVTTTTGIRR68 pKa = 11.84DD69 pKa = 4.01LQTLLLGSLSLLCAACSRR87 pKa = 11.84DD88 pKa = 3.36AFCNRR93 pKa = 11.84RR94 pKa = 11.84PQRR97 pKa = 11.84NGFF100 pKa = 3.55
MM1 pKa = 7.66KK2 pKa = 10.38GQDD5 pKa = 3.51VVLQDD10 pKa = 4.08LASEE14 pKa = 4.65LEE16 pKa = 4.95DD17 pKa = 3.08IVSPINLDD25 pKa = 3.47CEE27 pKa = 4.39EE28 pKa = 4.55EE29 pKa = 4.24IEE31 pKa = 4.29TEE33 pKa = 4.04EE34 pKa = 5.12VEE36 pKa = 4.47VDD38 pKa = 4.11FPNPYY43 pKa = 10.16EE44 pKa = 4.09ITAVCYY50 pKa = 10.43VCEE53 pKa = 4.03EE54 pKa = 4.21TLRR57 pKa = 11.84IAVVTTTTGIRR68 pKa = 11.84DD69 pKa = 4.01LQTLLLGSLSLLCAACSRR87 pKa = 11.84DD88 pKa = 3.36AFCNRR93 pKa = 11.84RR94 pKa = 11.84PQRR97 pKa = 11.84NGFF100 pKa = 3.55
Molecular weight: 11.14 kDa
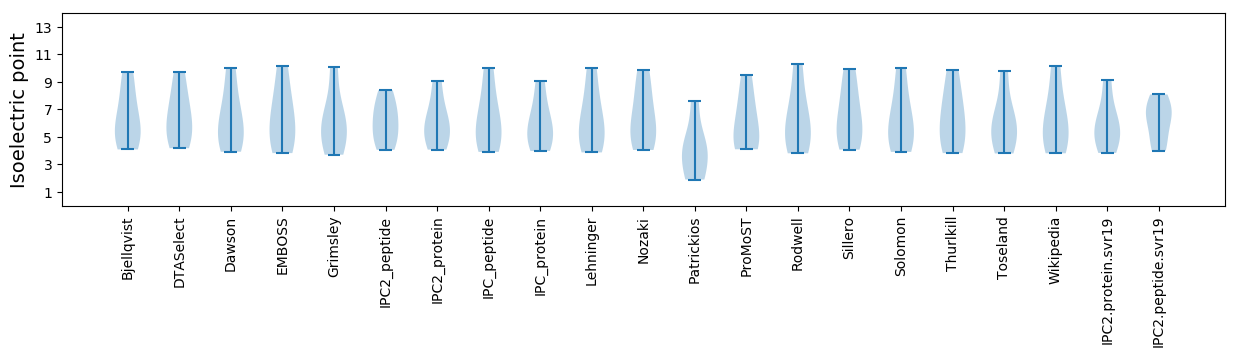
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5JS20|A0A0B5JS20_BPV3 Minor capsid protein L2 OS=Bos taurus papillomavirus 15 OX=1969700 GN=L2 PE=3 SV=1
MM1 pKa = 8.14DD2 pKa = 4.43SLEE5 pKa = 4.14ARR7 pKa = 11.84FDD9 pKa = 3.76ALQDD13 pKa = 3.41QLLEE17 pKa = 4.52IIEE20 pKa = 4.46RR21 pKa = 11.84DD22 pKa = 3.73SNTLEE27 pKa = 3.85VAIDD31 pKa = 3.46YY32 pKa = 7.97WSLLRR37 pKa = 11.84KK38 pKa = 9.53EE39 pKa = 4.07NAFYY43 pKa = 11.11YY44 pKa = 8.76HH45 pKa = 6.94ARR47 pKa = 11.84MQGKK51 pKa = 7.27TRR53 pKa = 11.84LGLYY57 pKa = 7.31TVPTARR63 pKa = 11.84VAEE66 pKa = 4.66HH67 pKa = 6.64KK68 pKa = 10.61AKK70 pKa = 10.38EE71 pKa = 4.07AIKK74 pKa = 10.25ISLYY78 pKa = 10.73LKK80 pKa = 10.21SLQKK84 pKa = 10.58SEE86 pKa = 4.09FANLRR91 pKa = 11.84WSLVDD96 pKa = 3.31TSSEE100 pKa = 4.04NFSARR105 pKa = 11.84PEE107 pKa = 3.91NTFKK111 pKa = 11.11KK112 pKa = 9.81KK113 pKa = 9.5GQHH116 pKa = 3.94VTVIYY121 pKa = 10.81DD122 pKa = 3.41SDD124 pKa = 4.39SNNSMIYY131 pKa = 8.23TAWGEE136 pKa = 3.73IFYY139 pKa = 10.93VDD141 pKa = 5.86EE142 pKa = 4.36EE143 pKa = 4.67DD144 pKa = 3.27TWHH147 pKa = 6.88KK148 pKa = 10.95AKK150 pKa = 10.83SDD152 pKa = 3.21VDD154 pKa = 3.77YY155 pKa = 11.6DD156 pKa = 4.21GISYY160 pKa = 10.09TDD162 pKa = 3.51YY163 pKa = 10.6LGEE166 pKa = 3.75RR167 pKa = 11.84HH168 pKa = 6.1VYY170 pKa = 10.62VNFHH174 pKa = 7.17DD175 pKa = 4.41DD176 pKa = 3.17ARR178 pKa = 11.84LYY180 pKa = 9.69STLGQWEE187 pKa = 4.19VHH189 pKa = 5.83FEE191 pKa = 4.2NKK193 pKa = 9.46VLSPPVTSSIPPGPTDD209 pKa = 3.13RR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84PAEE215 pKa = 4.22TGQHH219 pKa = 5.82PSGLKK224 pKa = 9.45TGLTARR230 pKa = 11.84KK231 pKa = 9.77SSITHH236 pKa = 5.82SRR238 pKa = 11.84DD239 pKa = 2.54SRR241 pKa = 11.84QRR243 pKa = 11.84SRR245 pKa = 11.84SHH247 pKa = 5.28SRR249 pKa = 11.84SRR251 pKa = 11.84SRR253 pKa = 11.84SRR255 pKa = 11.84SPTKK259 pKa = 10.55RR260 pKa = 11.84PDD262 pKa = 3.0SRR264 pKa = 11.84GRR266 pKa = 11.84GTEE269 pKa = 3.66LQGPRR274 pKa = 11.84RR275 pKa = 11.84GRR277 pKa = 11.84GGGGGGGDD285 pKa = 3.86FGEE288 pKa = 4.94PSPPSPGQVGEE299 pKa = 4.13RR300 pKa = 11.84HH301 pKa = 6.14RR302 pKa = 11.84SPSRR306 pKa = 11.84KK307 pKa = 8.6SASRR311 pKa = 11.84VAQLIEE317 pKa = 4.04EE318 pKa = 4.54AKK320 pKa = 10.7DD321 pKa = 3.55PPVLLLEE328 pKa = 4.64GGANILKK335 pKa = 9.99CFRR338 pKa = 11.84RR339 pKa = 11.84RR340 pKa = 11.84CTQTHH345 pKa = 5.17PHH347 pKa = 7.12RR348 pKa = 11.84FQCMSTSWTWVSKK361 pKa = 9.55TSTVKK366 pKa = 10.15SRR368 pKa = 11.84HH369 pKa = 5.14RR370 pKa = 11.84MLVAFTNCDD379 pKa = 3.06QRR381 pKa = 11.84KK382 pKa = 8.93GFLLNVRR389 pKa = 11.84LPKK392 pKa = 10.67GVTAVKK398 pKa = 10.65GCLDD402 pKa = 3.55SLL404 pKa = 4.17
MM1 pKa = 8.14DD2 pKa = 4.43SLEE5 pKa = 4.14ARR7 pKa = 11.84FDD9 pKa = 3.76ALQDD13 pKa = 3.41QLLEE17 pKa = 4.52IIEE20 pKa = 4.46RR21 pKa = 11.84DD22 pKa = 3.73SNTLEE27 pKa = 3.85VAIDD31 pKa = 3.46YY32 pKa = 7.97WSLLRR37 pKa = 11.84KK38 pKa = 9.53EE39 pKa = 4.07NAFYY43 pKa = 11.11YY44 pKa = 8.76HH45 pKa = 6.94ARR47 pKa = 11.84MQGKK51 pKa = 7.27TRR53 pKa = 11.84LGLYY57 pKa = 7.31TVPTARR63 pKa = 11.84VAEE66 pKa = 4.66HH67 pKa = 6.64KK68 pKa = 10.61AKK70 pKa = 10.38EE71 pKa = 4.07AIKK74 pKa = 10.25ISLYY78 pKa = 10.73LKK80 pKa = 10.21SLQKK84 pKa = 10.58SEE86 pKa = 4.09FANLRR91 pKa = 11.84WSLVDD96 pKa = 3.31TSSEE100 pKa = 4.04NFSARR105 pKa = 11.84PEE107 pKa = 3.91NTFKK111 pKa = 11.11KK112 pKa = 9.81KK113 pKa = 9.5GQHH116 pKa = 3.94VTVIYY121 pKa = 10.81DD122 pKa = 3.41SDD124 pKa = 4.39SNNSMIYY131 pKa = 8.23TAWGEE136 pKa = 3.73IFYY139 pKa = 10.93VDD141 pKa = 5.86EE142 pKa = 4.36EE143 pKa = 4.67DD144 pKa = 3.27TWHH147 pKa = 6.88KK148 pKa = 10.95AKK150 pKa = 10.83SDD152 pKa = 3.21VDD154 pKa = 3.77YY155 pKa = 11.6DD156 pKa = 4.21GISYY160 pKa = 10.09TDD162 pKa = 3.51YY163 pKa = 10.6LGEE166 pKa = 3.75RR167 pKa = 11.84HH168 pKa = 6.1VYY170 pKa = 10.62VNFHH174 pKa = 7.17DD175 pKa = 4.41DD176 pKa = 3.17ARR178 pKa = 11.84LYY180 pKa = 9.69STLGQWEE187 pKa = 4.19VHH189 pKa = 5.83FEE191 pKa = 4.2NKK193 pKa = 9.46VLSPPVTSSIPPGPTDD209 pKa = 3.13RR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84PAEE215 pKa = 4.22TGQHH219 pKa = 5.82PSGLKK224 pKa = 9.45TGLTARR230 pKa = 11.84KK231 pKa = 9.77SSITHH236 pKa = 5.82SRR238 pKa = 11.84DD239 pKa = 2.54SRR241 pKa = 11.84QRR243 pKa = 11.84SRR245 pKa = 11.84SHH247 pKa = 5.28SRR249 pKa = 11.84SRR251 pKa = 11.84SRR253 pKa = 11.84SRR255 pKa = 11.84SPTKK259 pKa = 10.55RR260 pKa = 11.84PDD262 pKa = 3.0SRR264 pKa = 11.84GRR266 pKa = 11.84GTEE269 pKa = 3.66LQGPRR274 pKa = 11.84RR275 pKa = 11.84GRR277 pKa = 11.84GGGGGGGDD285 pKa = 3.86FGEE288 pKa = 4.94PSPPSPGQVGEE299 pKa = 4.13RR300 pKa = 11.84HH301 pKa = 6.14RR302 pKa = 11.84SPSRR306 pKa = 11.84KK307 pKa = 8.6SASRR311 pKa = 11.84VAQLIEE317 pKa = 4.04EE318 pKa = 4.54AKK320 pKa = 10.7DD321 pKa = 3.55PPVLLLEE328 pKa = 4.64GGANILKK335 pKa = 9.99CFRR338 pKa = 11.84RR339 pKa = 11.84RR340 pKa = 11.84CTQTHH345 pKa = 5.17PHH347 pKa = 7.12RR348 pKa = 11.84FQCMSTSWTWVSKK361 pKa = 9.55TSTVKK366 pKa = 10.15SRR368 pKa = 11.84HH369 pKa = 5.14RR370 pKa = 11.84MLVAFTNCDD379 pKa = 3.06QRR381 pKa = 11.84KK382 pKa = 8.93GFLLNVRR389 pKa = 11.84LPKK392 pKa = 10.67GVTAVKK398 pKa = 10.65GCLDD402 pKa = 3.55SLL404 pKa = 4.17
Molecular weight: 45.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2180 |
42 |
609 |
363.3 |
40.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.367 ± 0.634 | 1.972 ± 0.618 |
5.963 ± 0.368 | 6.468 ± 0.415 |
5.0 ± 0.428 | 6.789 ± 0.665 |
2.156 ± 0.32 | 4.817 ± 0.655 |
5.046 ± 0.92 | 8.44 ± 1.654 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.743 ± 0.302 | 4.817 ± 0.704 |
5.963 ± 1.164 | 4.312 ± 0.428 |
6.33 ± 0.751 | 7.936 ± 0.714 |
5.963 ± 0.419 | 6.468 ± 0.639 |
1.514 ± 0.312 | 2.936 ± 0.261 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
