
Siphonobacter aquaeclarae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Siphonobacter
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
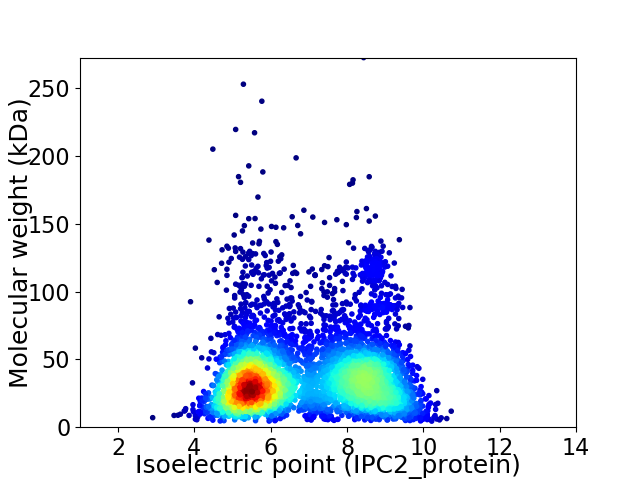
Virtual 2D-PAGE plot for 4964 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9SIY5|A0A1G9SIY5_9BACT Type III pantothenate kinase OS=Siphonobacter aquaeclarae OX=563176 GN=coaX PE=3 SV=1
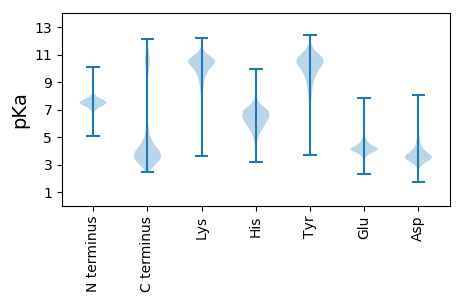
MM1 pKa = 7.58KK2 pKa = 10.35LFRR5 pKa = 11.84YY6 pKa = 7.84VTEE9 pKa = 4.05EE10 pKa = 3.85DD11 pKa = 4.26VIPGILIDD19 pKa = 4.17GKK21 pKa = 9.77PYY23 pKa = 9.84SLEE26 pKa = 3.95GFFEE30 pKa = 4.6AFGEE34 pKa = 4.27EE35 pKa = 3.81TGAYY39 pKa = 10.23DD40 pKa = 3.52LDD42 pKa = 3.77EE43 pKa = 4.5YY44 pKa = 11.18FFEE47 pKa = 4.63EE48 pKa = 3.95EE49 pKa = 4.52GIAVLEE55 pKa = 5.17DD56 pKa = 3.46YY57 pKa = 11.06LEE59 pKa = 4.24QYY61 pKa = 7.54GTSEE65 pKa = 5.14LEE67 pKa = 4.19PLDD70 pKa = 5.2EE71 pKa = 5.62DD72 pKa = 3.41IQYY75 pKa = 10.3ASCVARR81 pKa = 11.84PSKK84 pKa = 10.33IICIGLNYY92 pKa = 9.74ADD94 pKa = 4.19HH95 pKa = 6.94AKK97 pKa = 8.59EE98 pKa = 3.88TGAPIPQEE106 pKa = 3.73PIIFFKK112 pKa = 10.58ATTALCGPNDD122 pKa = 3.79DD123 pKa = 4.28VVIPRR128 pKa = 11.84NSVKK132 pKa = 10.08TDD134 pKa = 3.12WEE136 pKa = 4.2VEE138 pKa = 4.01LAVVIGAEE146 pKa = 3.99CSYY149 pKa = 9.7VTEE152 pKa = 4.29EE153 pKa = 4.26QAVDD157 pKa = 3.93YY158 pKa = 10.44IAGYY162 pKa = 11.1AVMNDD167 pKa = 3.12YY168 pKa = 11.04SEE170 pKa = 6.23RR171 pKa = 11.84EE172 pKa = 3.94FQLEE176 pKa = 5.49RR177 pKa = 11.84GGQWDD182 pKa = 3.74KK183 pKa = 11.9GKK185 pKa = 11.04GCDD188 pKa = 3.22TFGPLGPYY196 pKa = 10.06LVTPDD201 pKa = 4.01EE202 pKa = 4.43VDD204 pKa = 4.57DD205 pKa = 4.31VDD207 pKa = 5.95SLPMWLKK214 pKa = 10.91VNGKK218 pKa = 6.67TFQNSTTAQMIFKK231 pKa = 9.13VPQVISYY238 pKa = 9.64LSQFMTLLPGDD249 pKa = 4.39VISTGTPPGVGLGFNPPIYY268 pKa = 10.07LKK270 pKa = 10.69PGDD273 pKa = 3.93VVEE276 pKa = 5.18LGIEE280 pKa = 4.12GLGEE284 pKa = 3.81QRR286 pKa = 11.84QVAVSWEE293 pKa = 4.03DD294 pKa = 3.34ANN296 pKa = 4.42
MM1 pKa = 7.58KK2 pKa = 10.35LFRR5 pKa = 11.84YY6 pKa = 7.84VTEE9 pKa = 4.05EE10 pKa = 3.85DD11 pKa = 4.26VIPGILIDD19 pKa = 4.17GKK21 pKa = 9.77PYY23 pKa = 9.84SLEE26 pKa = 3.95GFFEE30 pKa = 4.6AFGEE34 pKa = 4.27EE35 pKa = 3.81TGAYY39 pKa = 10.23DD40 pKa = 3.52LDD42 pKa = 3.77EE43 pKa = 4.5YY44 pKa = 11.18FFEE47 pKa = 4.63EE48 pKa = 3.95EE49 pKa = 4.52GIAVLEE55 pKa = 5.17DD56 pKa = 3.46YY57 pKa = 11.06LEE59 pKa = 4.24QYY61 pKa = 7.54GTSEE65 pKa = 5.14LEE67 pKa = 4.19PLDD70 pKa = 5.2EE71 pKa = 5.62DD72 pKa = 3.41IQYY75 pKa = 10.3ASCVARR81 pKa = 11.84PSKK84 pKa = 10.33IICIGLNYY92 pKa = 9.74ADD94 pKa = 4.19HH95 pKa = 6.94AKK97 pKa = 8.59EE98 pKa = 3.88TGAPIPQEE106 pKa = 3.73PIIFFKK112 pKa = 10.58ATTALCGPNDD122 pKa = 3.79DD123 pKa = 4.28VVIPRR128 pKa = 11.84NSVKK132 pKa = 10.08TDD134 pKa = 3.12WEE136 pKa = 4.2VEE138 pKa = 4.01LAVVIGAEE146 pKa = 3.99CSYY149 pKa = 9.7VTEE152 pKa = 4.29EE153 pKa = 4.26QAVDD157 pKa = 3.93YY158 pKa = 10.44IAGYY162 pKa = 11.1AVMNDD167 pKa = 3.12YY168 pKa = 11.04SEE170 pKa = 6.23RR171 pKa = 11.84EE172 pKa = 3.94FQLEE176 pKa = 5.49RR177 pKa = 11.84GGQWDD182 pKa = 3.74KK183 pKa = 11.9GKK185 pKa = 11.04GCDD188 pKa = 3.22TFGPLGPYY196 pKa = 10.06LVTPDD201 pKa = 4.01EE202 pKa = 4.43VDD204 pKa = 4.57DD205 pKa = 4.31VDD207 pKa = 5.95SLPMWLKK214 pKa = 10.91VNGKK218 pKa = 6.67TFQNSTTAQMIFKK231 pKa = 9.13VPQVISYY238 pKa = 9.64LSQFMTLLPGDD249 pKa = 4.39VISTGTPPGVGLGFNPPIYY268 pKa = 10.07LKK270 pKa = 10.69PGDD273 pKa = 3.93VVEE276 pKa = 5.18LGIEE280 pKa = 4.12GLGEE284 pKa = 3.81QRR286 pKa = 11.84QVAVSWEE293 pKa = 4.03DD294 pKa = 3.34ANN296 pKa = 4.42
Molecular weight: 32.71 kDa
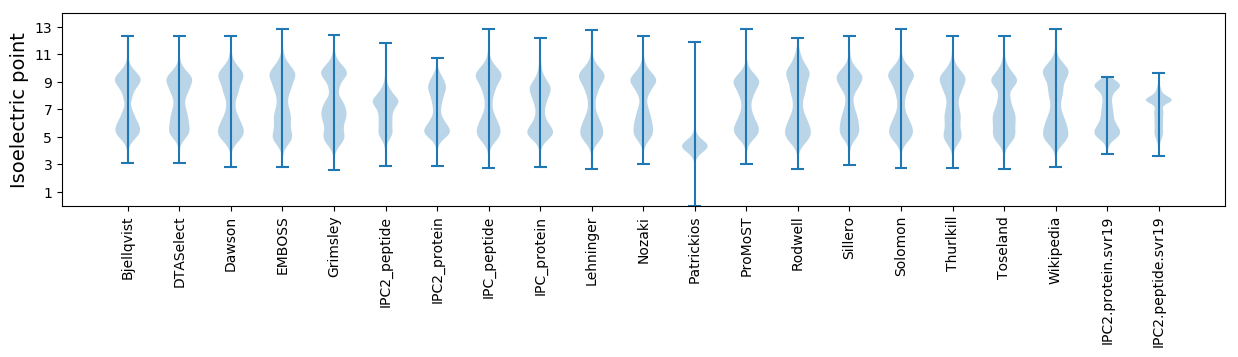
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9RIP6|A0A1G9RIP6_9BACT Uncharacterized protein OS=Siphonobacter aquaeclarae OX=563176 GN=SAMN04488090_2901 PE=4 SV=1
MM1 pKa = 7.34KK2 pKa = 10.23RR3 pKa = 11.84LSAFFLFLTLSASAQTPEE21 pKa = 3.08QWYY24 pKa = 9.89RR25 pKa = 11.84RR26 pKa = 11.84EE27 pKa = 3.97TGSRR31 pKa = 11.84LSSPNRR37 pKa = 11.84HH38 pKa = 5.9MLPDD42 pKa = 3.69SLFSGAFFRR51 pKa = 11.84CRR53 pKa = 11.84PLPAAHH59 pKa = 7.26PSLMAGASRR68 pKa = 11.84LACLSDD74 pKa = 3.85EE75 pKa = 4.57LSAIHH80 pKa = 5.83TRR82 pKa = 11.84IVQAQGEE89 pKa = 4.55TGNQIRR95 pKa = 11.84LAKK98 pKa = 10.45LLLDD102 pKa = 3.72VRR104 pKa = 11.84RR105 pKa = 11.84QAVEE109 pKa = 3.68QYY111 pKa = 11.56ALLRR115 pKa = 11.84NAAKK119 pKa = 10.46RR120 pKa = 11.84LPLVRR125 pKa = 11.84KK126 pKa = 9.67RR127 pKa = 11.84LNTYY131 pKa = 10.08VFDD134 pKa = 4.62GSCALTRR141 pKa = 11.84YY142 pKa = 10.09DD143 pKa = 4.46GLITEE148 pKa = 5.37LLGTAPPPIVPP159 pKa = 4.23
MM1 pKa = 7.34KK2 pKa = 10.23RR3 pKa = 11.84LSAFFLFLTLSASAQTPEE21 pKa = 3.08QWYY24 pKa = 9.89RR25 pKa = 11.84RR26 pKa = 11.84EE27 pKa = 3.97TGSRR31 pKa = 11.84LSSPNRR37 pKa = 11.84HH38 pKa = 5.9MLPDD42 pKa = 3.69SLFSGAFFRR51 pKa = 11.84CRR53 pKa = 11.84PLPAAHH59 pKa = 7.26PSLMAGASRR68 pKa = 11.84LACLSDD74 pKa = 3.85EE75 pKa = 4.57LSAIHH80 pKa = 5.83TRR82 pKa = 11.84IVQAQGEE89 pKa = 4.55TGNQIRR95 pKa = 11.84LAKK98 pKa = 10.45LLLDD102 pKa = 3.72VRR104 pKa = 11.84RR105 pKa = 11.84QAVEE109 pKa = 3.68QYY111 pKa = 11.56ALLRR115 pKa = 11.84NAAKK119 pKa = 10.46RR120 pKa = 11.84LPLVRR125 pKa = 11.84KK126 pKa = 9.67RR127 pKa = 11.84LNTYY131 pKa = 10.08VFDD134 pKa = 4.62GSCALTRR141 pKa = 11.84YY142 pKa = 10.09DD143 pKa = 4.46GLITEE148 pKa = 5.37LLGTAPPPIVPP159 pKa = 4.23
Molecular weight: 17.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1846786 |
38 |
2407 |
372.0 |
41.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.462 ± 0.033 | 0.724 ± 0.009 |
5.218 ± 0.024 | 5.704 ± 0.037 |
4.776 ± 0.024 | 7.671 ± 0.033 |
1.852 ± 0.017 | 5.464 ± 0.027 |
5.136 ± 0.03 | 10.095 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.976 ± 0.013 | 4.238 ± 0.033 |
4.621 ± 0.021 | 3.791 ± 0.018 |
5.758 ± 0.029 | 6.223 ± 0.03 |
6.004 ± 0.031 | 6.979 ± 0.027 |
1.45 ± 0.013 | 3.857 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
