
Parvibaculum sedimenti
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Parvibaculaceae; Parvibaculum
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
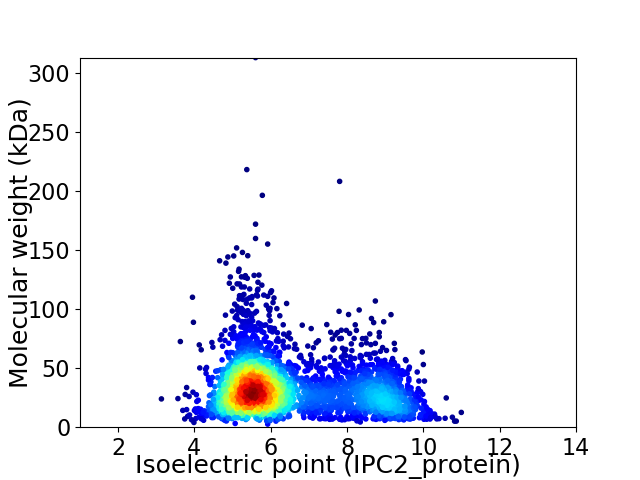
Virtual 2D-PAGE plot for 3429 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N6VMB7|A0A6N6VMB7_9RHIZ EamA family transporter OS=Parvibaculum sedimenti OX=2608632 GN=F2P47_09135 PE=4 SV=1
MM1 pKa = 7.9PYY3 pKa = 10.44SPVHH7 pKa = 4.89VRR9 pKa = 11.84RR10 pKa = 11.84SQRR13 pKa = 11.84RR14 pKa = 11.84ATKK17 pKa = 10.15LCALILPLSLWSGLAGADD35 pKa = 3.56TTISSSTTAQTTSADD50 pKa = 3.06GDD52 pKa = 3.74ITVTGTLSVGADD64 pKa = 3.23TAVTIDD70 pKa = 3.88SDD72 pKa = 4.14NSFTNEE78 pKa = 3.4GSTLAASAGGIGVSVTTATTGSIANTGTLRR108 pKa = 11.84TGTAASSDD116 pKa = 3.71TADD119 pKa = 4.33DD120 pKa = 4.27GVIGGPAIEE129 pKa = 5.0VYY131 pKa = 10.36QSLGGGISNAAGYY144 pKa = 8.09TITTYY149 pKa = 11.35GNNTIFVGTDD159 pKa = 2.76ATSPANITIGNVGTDD174 pKa = 3.17AEE176 pKa = 4.69AYY178 pKa = 9.74GIYY181 pKa = 9.79NAGYY185 pKa = 9.14IYY187 pKa = 10.08TYY189 pKa = 10.74GSNSGEE195 pKa = 3.8ATTAIRR201 pKa = 11.84VEE203 pKa = 4.11ADD205 pKa = 2.69SGYY208 pKa = 9.6TSDD211 pKa = 3.65LTGGISNAGTSSIILSRR228 pKa = 11.84AIDD231 pKa = 3.72ADD233 pKa = 3.43ATGISIGSGGIVDD246 pKa = 4.86EE247 pKa = 4.57IVNEE251 pKa = 4.26GTLEE255 pKa = 4.09ALTLGTTGGKK265 pKa = 9.58AVAISVEE272 pKa = 4.09AGGTLNGITNSGTIEE287 pKa = 3.98ASASTVGDD295 pKa = 3.24AVAIVDD301 pKa = 3.97ASGTLSSIVNSGTIEE316 pKa = 4.62AIANDD321 pKa = 4.26GAATAIDD328 pKa = 3.95VSTSASAFTLTNSGTITGAIYY349 pKa = 10.45LGSGASTITSDD360 pKa = 5.01DD361 pKa = 3.33GTITGNIDD369 pKa = 3.02IASGGTLDD377 pKa = 3.78MSLSGGAEE385 pKa = 3.8FTGGTNTGATGTLDD399 pKa = 3.48VQDD402 pKa = 3.51ATFRR406 pKa = 11.84SAGSGYY412 pKa = 10.61YY413 pKa = 9.12ATTAAFASDD422 pKa = 3.93SIFAVTYY429 pKa = 10.25DD430 pKa = 3.45AVANTSALLTTTGTTSFAAGATVDD454 pKa = 3.79VTFSSYY460 pKa = 11.17LSTSEE465 pKa = 4.28TINLVDD471 pKa = 5.05AGTLSLSAGGVDD483 pKa = 4.2DD484 pKa = 5.22LLIGGVSAGYY494 pKa = 9.84NAVLSTSGNEE504 pKa = 4.22LYY506 pKa = 9.08ITLARR511 pKa = 11.84KK512 pKa = 7.1TAAEE516 pKa = 4.1LGITGNAATIYY527 pKa = 10.31NVAPTAFALDD537 pKa = 4.18DD538 pKa = 3.92EE539 pKa = 5.27FGAAVGNLSTVAAIQKK555 pKa = 9.76LYY557 pKa = 11.02KK558 pKa = 10.41DD559 pKa = 4.67LLPDD563 pKa = 3.72LSGARR568 pKa = 11.84EE569 pKa = 3.9EE570 pKa = 4.19QALRR574 pKa = 11.84VQDD577 pKa = 3.44ISSGIVSSRR586 pKa = 11.84LDD588 pKa = 3.61LLRR591 pKa = 11.84SDD593 pKa = 5.03DD594 pKa = 4.4QDD596 pKa = 3.68GQTTGYY602 pKa = 9.66RR603 pKa = 11.84RR604 pKa = 11.84RR605 pKa = 11.84RR606 pKa = 11.84NTGLWAQEE614 pKa = 3.83AVSAEE619 pKa = 4.11SGAGGEE625 pKa = 4.54KK626 pKa = 10.02SASYY630 pKa = 11.02DD631 pKa = 3.22GTLYY635 pKa = 11.26ALAVGYY641 pKa = 7.48DD642 pKa = 3.38TRR644 pKa = 11.84DD645 pKa = 2.95GDD647 pKa = 4.11GDD649 pKa = 3.67VSGASFTYY657 pKa = 10.75AAMQYY662 pKa = 10.02GASDD666 pKa = 3.73PAHH669 pKa = 7.13DD670 pKa = 4.85NVNQTYY676 pKa = 10.93LLQLYY681 pKa = 9.18HH682 pKa = 6.88ALNRR686 pKa = 11.84GAFYY690 pKa = 10.15WDD692 pKa = 3.16AMGSLGLDD700 pKa = 3.11SYY702 pKa = 11.26KK703 pKa = 10.74GYY705 pKa = 10.37RR706 pKa = 11.84QVTAGDD712 pKa = 3.69VTRR715 pKa = 11.84NPYY718 pKa = 9.64ANWMGYY724 pKa = 7.31QAGLSTQVGYY734 pKa = 10.9DD735 pKa = 3.55LALGPISIRR744 pKa = 11.84PALGASYY751 pKa = 9.5TYY753 pKa = 10.69LRR755 pKa = 11.84QASYY759 pKa = 10.45TEE761 pKa = 4.06EE762 pKa = 4.93GGGDD766 pKa = 3.81GVDD769 pKa = 3.68LAINANSFQSFRR781 pKa = 11.84TTAEE785 pKa = 3.67LRR787 pKa = 11.84VSGKK791 pKa = 8.72TGGNPDD797 pKa = 3.27IDD799 pKa = 4.05PYY801 pKa = 11.29LRR803 pKa = 11.84GGITHH808 pKa = 7.22EE809 pKa = 4.63FLDD812 pKa = 4.31ATPVADD818 pKa = 3.49GHH820 pKa = 5.89FVAGGAFSVEE830 pKa = 3.68GDD832 pKa = 4.33ALDD835 pKa = 4.34KK836 pKa = 10.84DD837 pKa = 3.83IPFAGLGLGIGSGYY851 pKa = 10.93ARR853 pKa = 11.84LNFDD857 pKa = 3.23YY858 pKa = 10.53TGQFGDD864 pKa = 4.43KK865 pKa = 9.1LTSHH869 pKa = 6.46QATATFVMKK878 pKa = 10.16FF879 pKa = 3.22
MM1 pKa = 7.9PYY3 pKa = 10.44SPVHH7 pKa = 4.89VRR9 pKa = 11.84RR10 pKa = 11.84SQRR13 pKa = 11.84RR14 pKa = 11.84ATKK17 pKa = 10.15LCALILPLSLWSGLAGADD35 pKa = 3.56TTISSSTTAQTTSADD50 pKa = 3.06GDD52 pKa = 3.74ITVTGTLSVGADD64 pKa = 3.23TAVTIDD70 pKa = 3.88SDD72 pKa = 4.14NSFTNEE78 pKa = 3.4GSTLAASAGGIGVSVTTATTGSIANTGTLRR108 pKa = 11.84TGTAASSDD116 pKa = 3.71TADD119 pKa = 4.33DD120 pKa = 4.27GVIGGPAIEE129 pKa = 5.0VYY131 pKa = 10.36QSLGGGISNAAGYY144 pKa = 8.09TITTYY149 pKa = 11.35GNNTIFVGTDD159 pKa = 2.76ATSPANITIGNVGTDD174 pKa = 3.17AEE176 pKa = 4.69AYY178 pKa = 9.74GIYY181 pKa = 9.79NAGYY185 pKa = 9.14IYY187 pKa = 10.08TYY189 pKa = 10.74GSNSGEE195 pKa = 3.8ATTAIRR201 pKa = 11.84VEE203 pKa = 4.11ADD205 pKa = 2.69SGYY208 pKa = 9.6TSDD211 pKa = 3.65LTGGISNAGTSSIILSRR228 pKa = 11.84AIDD231 pKa = 3.72ADD233 pKa = 3.43ATGISIGSGGIVDD246 pKa = 4.86EE247 pKa = 4.57IVNEE251 pKa = 4.26GTLEE255 pKa = 4.09ALTLGTTGGKK265 pKa = 9.58AVAISVEE272 pKa = 4.09AGGTLNGITNSGTIEE287 pKa = 3.98ASASTVGDD295 pKa = 3.24AVAIVDD301 pKa = 3.97ASGTLSSIVNSGTIEE316 pKa = 4.62AIANDD321 pKa = 4.26GAATAIDD328 pKa = 3.95VSTSASAFTLTNSGTITGAIYY349 pKa = 10.45LGSGASTITSDD360 pKa = 5.01DD361 pKa = 3.33GTITGNIDD369 pKa = 3.02IASGGTLDD377 pKa = 3.78MSLSGGAEE385 pKa = 3.8FTGGTNTGATGTLDD399 pKa = 3.48VQDD402 pKa = 3.51ATFRR406 pKa = 11.84SAGSGYY412 pKa = 10.61YY413 pKa = 9.12ATTAAFASDD422 pKa = 3.93SIFAVTYY429 pKa = 10.25DD430 pKa = 3.45AVANTSALLTTTGTTSFAAGATVDD454 pKa = 3.79VTFSSYY460 pKa = 11.17LSTSEE465 pKa = 4.28TINLVDD471 pKa = 5.05AGTLSLSAGGVDD483 pKa = 4.2DD484 pKa = 5.22LLIGGVSAGYY494 pKa = 9.84NAVLSTSGNEE504 pKa = 4.22LYY506 pKa = 9.08ITLARR511 pKa = 11.84KK512 pKa = 7.1TAAEE516 pKa = 4.1LGITGNAATIYY527 pKa = 10.31NVAPTAFALDD537 pKa = 4.18DD538 pKa = 3.92EE539 pKa = 5.27FGAAVGNLSTVAAIQKK555 pKa = 9.76LYY557 pKa = 11.02KK558 pKa = 10.41DD559 pKa = 4.67LLPDD563 pKa = 3.72LSGARR568 pKa = 11.84EE569 pKa = 3.9EE570 pKa = 4.19QALRR574 pKa = 11.84VQDD577 pKa = 3.44ISSGIVSSRR586 pKa = 11.84LDD588 pKa = 3.61LLRR591 pKa = 11.84SDD593 pKa = 5.03DD594 pKa = 4.4QDD596 pKa = 3.68GQTTGYY602 pKa = 9.66RR603 pKa = 11.84RR604 pKa = 11.84RR605 pKa = 11.84RR606 pKa = 11.84NTGLWAQEE614 pKa = 3.83AVSAEE619 pKa = 4.11SGAGGEE625 pKa = 4.54KK626 pKa = 10.02SASYY630 pKa = 11.02DD631 pKa = 3.22GTLYY635 pKa = 11.26ALAVGYY641 pKa = 7.48DD642 pKa = 3.38TRR644 pKa = 11.84DD645 pKa = 2.95GDD647 pKa = 4.11GDD649 pKa = 3.67VSGASFTYY657 pKa = 10.75AAMQYY662 pKa = 10.02GASDD666 pKa = 3.73PAHH669 pKa = 7.13DD670 pKa = 4.85NVNQTYY676 pKa = 10.93LLQLYY681 pKa = 9.18HH682 pKa = 6.88ALNRR686 pKa = 11.84GAFYY690 pKa = 10.15WDD692 pKa = 3.16AMGSLGLDD700 pKa = 3.11SYY702 pKa = 11.26KK703 pKa = 10.74GYY705 pKa = 10.37RR706 pKa = 11.84QVTAGDD712 pKa = 3.69VTRR715 pKa = 11.84NPYY718 pKa = 9.64ANWMGYY724 pKa = 7.31QAGLSTQVGYY734 pKa = 10.9DD735 pKa = 3.55LALGPISIRR744 pKa = 11.84PALGASYY751 pKa = 9.5TYY753 pKa = 10.69LRR755 pKa = 11.84QASYY759 pKa = 10.45TEE761 pKa = 4.06EE762 pKa = 4.93GGGDD766 pKa = 3.81GVDD769 pKa = 3.68LAINANSFQSFRR781 pKa = 11.84TTAEE785 pKa = 3.67LRR787 pKa = 11.84VSGKK791 pKa = 8.72TGGNPDD797 pKa = 3.27IDD799 pKa = 4.05PYY801 pKa = 11.29LRR803 pKa = 11.84GGITHH808 pKa = 7.22EE809 pKa = 4.63FLDD812 pKa = 4.31ATPVADD818 pKa = 3.49GHH820 pKa = 5.89FVAGGAFSVEE830 pKa = 3.68GDD832 pKa = 4.33ALDD835 pKa = 4.34KK836 pKa = 10.84DD837 pKa = 3.83IPFAGLGLGIGSGYY851 pKa = 10.93ARR853 pKa = 11.84LNFDD857 pKa = 3.23YY858 pKa = 10.53TGQFGDD864 pKa = 4.43KK865 pKa = 9.1LTSHH869 pKa = 6.46QATATFVMKK878 pKa = 10.16FF879 pKa = 3.22
Molecular weight: 88.77 kDa
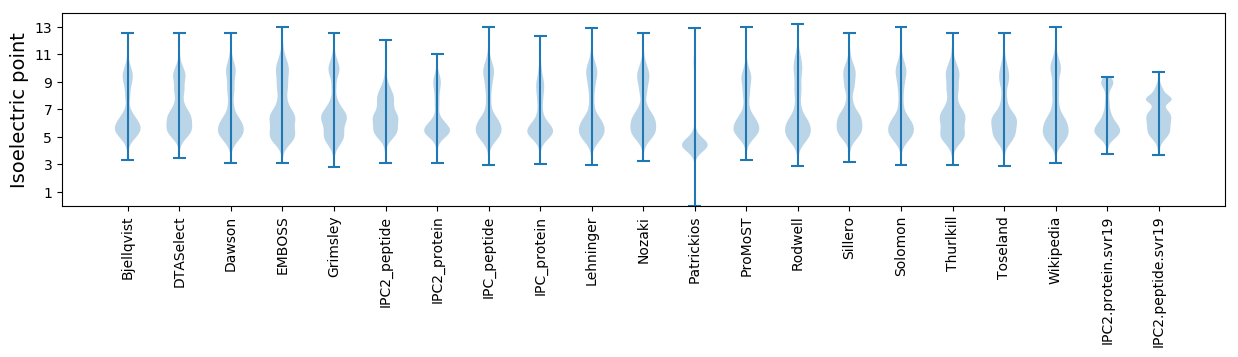
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N6VGA5|A0A6N6VGA5_9RHIZ Putative sulfate exporter family transporter OS=Parvibaculum sedimenti OX=2608632 GN=F2P47_12745 PE=3 SV=1
MM1 pKa = 7.49AFVRR5 pKa = 11.84FAGRR9 pKa = 11.84KK10 pKa = 4.7VTVPGSPWMRR20 pKa = 11.84KK21 pKa = 5.64TAGAALIGGGALGFLPVLGFWMIPLGVVVLSVDD54 pKa = 3.09SHH56 pKa = 5.34RR57 pKa = 11.84VRR59 pKa = 11.84RR60 pKa = 11.84VRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84TEE66 pKa = 3.65VWWGRR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84SS75 pKa = 3.07
MM1 pKa = 7.49AFVRR5 pKa = 11.84FAGRR9 pKa = 11.84KK10 pKa = 4.7VTVPGSPWMRR20 pKa = 11.84KK21 pKa = 5.64TAGAALIGGGALGFLPVLGFWMIPLGVVVLSVDD54 pKa = 3.09SHH56 pKa = 5.34RR57 pKa = 11.84VRR59 pKa = 11.84RR60 pKa = 11.84VRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84TEE66 pKa = 3.65VWWGRR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84SS75 pKa = 3.07
Molecular weight: 8.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1103409 |
25 |
2854 |
321.8 |
35.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.149 ± 0.063 | 0.844 ± 0.013 |
5.644 ± 0.028 | 6.183 ± 0.037 |
3.81 ± 0.027 | 8.503 ± 0.04 |
2.162 ± 0.016 | 5.333 ± 0.027 |
3.896 ± 0.039 | 9.909 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.49 ± 0.018 | 2.714 ± 0.024 |
5.075 ± 0.026 | 2.838 ± 0.023 |
6.956 ± 0.049 | 5.308 ± 0.031 |
5.186 ± 0.029 | 7.27 ± 0.035 |
1.315 ± 0.015 | 2.414 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
