
Macrostomum lignano
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Platyhelminthes; Rhabditophora; Macrostomorpha; Macrostomida; Macrostomidae; Macrostomum
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
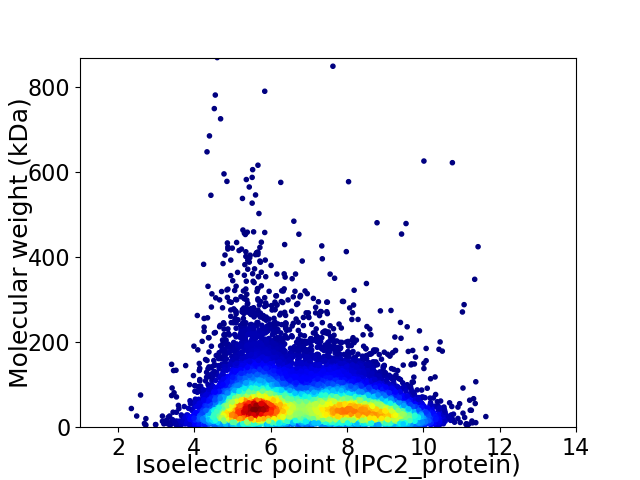
Virtual 2D-PAGE plot for 43466 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A267ETZ5|A0A267ETZ5_9PLAT Sulfotransfer_1 domain-containing protein OS=Macrostomum lignano OX=282301 GN=BOX15_Mlig003539g1 PE=4 SV=1
MM1 pKa = 7.62APNGVSDD8 pKa = 4.2RR9 pKa = 11.84SARR12 pKa = 11.84LPRR15 pKa = 11.84LIMIRR20 pKa = 11.84ILLLVAMQATLFPGTAFAQLAWDD43 pKa = 4.14TFSPAQQTYY52 pKa = 7.7PTATVTIDD60 pKa = 3.04EE61 pKa = 4.44NMAATTTVLTITTTGATGTVTYY83 pKa = 10.26SIASSTTAAFTIGASTGVITVNDD106 pKa = 3.66ASLLDD111 pKa = 3.71RR112 pKa = 11.84EE113 pKa = 4.88AISFIDD119 pKa = 4.29LVLQAVDD126 pKa = 3.62TSPATVSTTVRR137 pKa = 11.84IDD139 pKa = 3.46LKK141 pKa = 10.77DD142 pKa = 3.65VNEE145 pKa = 4.08IAPTFSSTSTYY156 pKa = 10.81SVTISEE162 pKa = 4.55SATVGTTIQALTVTDD177 pKa = 3.69TDD179 pKa = 3.87SAATNVACTISSGDD193 pKa = 3.72PDD195 pKa = 3.64GKK197 pKa = 10.71FQFRR201 pKa = 11.84SDD203 pKa = 3.34NQTIEE208 pKa = 4.14LKK210 pKa = 10.85AAIEE214 pKa = 4.18PDD216 pKa = 3.55SPTNNSLTYY225 pKa = 9.79TLTIVCYY232 pKa = 10.79DD233 pKa = 3.69NFNGVYY239 pKa = 10.14SGGSLASTGTVSVSVTPVNDD259 pKa = 3.98NSPTWGTFTPALATPITTIDD279 pKa = 3.5EE280 pKa = 4.24NLASGTIFTAAASDD294 pKa = 3.48VDD296 pKa = 4.09YY297 pKa = 8.96GTDD300 pKa = 3.47GVVTFSFSTVKK311 pKa = 10.6FNTGADD317 pKa = 3.29ASALFTIGSSSGVVSRR333 pKa = 11.84TAATTPALDD342 pKa = 3.46RR343 pKa = 11.84EE344 pKa = 4.89TYY346 pKa = 9.25SHH348 pKa = 6.83VEE350 pKa = 4.04FVVLAMDD357 pKa = 4.43SPTVTADD364 pKa = 3.5RR365 pKa = 11.84LTITQTVQVNLKK377 pKa = 9.81DD378 pKa = 3.91VNEE381 pKa = 4.15FTPVFDD387 pKa = 5.16PSTGLYY393 pKa = 9.52NVSFSEE399 pKa = 4.21TAPAGTLVLNVSVSDD414 pKa = 3.9PDD416 pKa = 3.99FSRR419 pKa = 11.84TAISCTITSGNTLDD433 pKa = 3.9RR434 pKa = 11.84FQMRR438 pKa = 11.84ADD440 pKa = 3.71GTSLEE445 pKa = 4.92LKK447 pKa = 8.65TQIEE451 pKa = 4.42PDD453 pKa = 3.93SPTSDD458 pKa = 4.1PINYY462 pKa = 9.36VLTLSCADD470 pKa = 3.66NTAGTYY476 pKa = 10.31SGGQKK481 pKa = 10.3SSTGTVTVWVEE492 pKa = 3.66YY493 pKa = 11.02NNDD496 pKa = 3.57YY497 pKa = 11.24SPVWSTFSPAWTSATTPISTILEE520 pKa = 4.01NLAVNQTVFTASATDD535 pKa = 4.34DD536 pKa = 3.9DD537 pKa = 5.62AGDD540 pKa = 4.65DD541 pKa = 3.93GLVVYY546 pKa = 10.13SISGAVAEE554 pKa = 4.69NGSSVMEE561 pKa = 4.03AFAINGSSGAVSIANSTVLDD581 pKa = 3.81RR582 pKa = 11.84EE583 pKa = 4.82GYY585 pKa = 9.46SYY587 pKa = 12.0VDD589 pKa = 3.35VTVEE593 pKa = 4.26ARR595 pKa = 11.84DD596 pKa = 3.9SPASGTTYY604 pKa = 10.46TITQTVRR611 pKa = 11.84IGISDD616 pKa = 3.83YY617 pKa = 11.79NEE619 pKa = 3.95FPPAFTPSTGLYY631 pKa = 8.94TPAIPEE637 pKa = 4.06TTTAGTAVQTIVATDD652 pKa = 3.64PDD654 pKa = 4.28YY655 pKa = 11.85SNTAVVCAIAAGNSLNRR672 pKa = 11.84FAINTADD679 pKa = 3.58KK680 pKa = 10.83KK681 pKa = 10.75LVEE684 pKa = 4.42TASVIQPDD692 pKa = 4.8KK693 pKa = 9.98PTLDD697 pKa = 3.3PTIYY701 pKa = 10.86LLTVHH706 pKa = 6.63CADD709 pKa = 4.41MNNGTLGTPVLTSTAYY725 pKa = 8.23ITVNVTLVNDD735 pKa = 3.79YY736 pKa = 11.47DD737 pKa = 3.76PTITLNSTTMVIPEE751 pKa = 4.35DD752 pKa = 4.05TPVSSFIVSVTADD765 pKa = 3.62DD766 pKa = 4.94LDD768 pKa = 4.08YY769 pKa = 11.75GNDD772 pKa = 3.55GTFTISLSPTTPFALTGDD790 pKa = 3.83RR791 pKa = 11.84TKK793 pKa = 11.31LLLTEE798 pKa = 4.29ALDD801 pKa = 4.37RR802 pKa = 11.84EE803 pKa = 4.73TKK805 pKa = 10.12DD806 pKa = 3.04SYY808 pKa = 10.22VIEE811 pKa = 4.26MRR813 pKa = 11.84ALDD816 pKa = 3.63QGSPARR822 pKa = 11.84SSTTTLTVSITDD834 pKa = 3.62VNDD837 pKa = 3.28NTPVCSPNTYY847 pKa = 9.75NVSVTADD854 pKa = 3.18SAATLVASIPCTDD867 pKa = 4.48ADD869 pKa = 4.88LGDD872 pKa = 4.12NQLLAYY878 pKa = 10.13SFLSGDD884 pKa = 3.55TRR886 pKa = 11.84NLTLSSTTGALTTTFLTYY904 pKa = 10.88DD905 pKa = 3.13SAAPSKK911 pKa = 9.64TLVITVRR918 pKa = 11.84DD919 pKa = 3.69SATPASAQRR928 pKa = 11.84TATVTVTVHH937 pKa = 5.35VLPRR941 pKa = 11.84NLGPPVWGTFNPPWISATTAINSVSEE967 pKa = 4.06NADD970 pKa = 2.94EE971 pKa = 4.6GFAVFTAVAIDD982 pKa = 4.18YY983 pKa = 10.81DD984 pKa = 4.28SGPDD988 pKa = 3.54GQVIYY993 pKa = 10.93SLASVTGTDD1002 pKa = 3.25GTTSVDD1008 pKa = 3.27LSSVFNISSSSGQVQVKK1025 pKa = 8.89TKK1027 pKa = 10.87GILDD1031 pKa = 3.97RR1032 pKa = 11.84EE1033 pKa = 4.3ATPAYY1038 pKa = 9.14TYY1040 pKa = 11.01INFVVQARR1048 pKa = 11.84DD1049 pKa = 3.34NPTTGTPNSIIQTVRR1064 pKa = 11.84VDD1066 pKa = 3.32VTDD1069 pKa = 4.15FNEE1072 pKa = 4.44LDD1074 pKa = 3.52PTFTGDD1080 pKa = 3.48PFSATLVEE1088 pKa = 5.13DD1089 pKa = 3.87SAAGTLVKK1097 pKa = 10.7QMVVSDD1103 pKa = 4.42PDD1105 pKa = 3.46SSNTIITCQITSGNDD1120 pKa = 2.96LSLFRR1125 pKa = 11.84TNSANSTIIEE1135 pKa = 4.82LNTAIDD1141 pKa = 3.56PDD1143 pKa = 3.76RR1144 pKa = 11.84PKK1146 pKa = 10.92NHH1148 pKa = 7.07PLSYY1152 pKa = 10.88LLIVTCLDD1160 pKa = 3.34NNGAGSYY1167 pKa = 7.87RR1168 pKa = 11.84TATANLSVTITTTNDD1183 pKa = 2.93HH1184 pKa = 6.35TPVVSIPIGSSITLPEE1200 pKa = 3.84TQAIGDD1206 pKa = 3.77VADD1209 pKa = 3.98ISVVDD1214 pKa = 3.63QDD1216 pKa = 3.98YY1217 pKa = 8.57GTDD1220 pKa = 3.48GTVACSIAPTGTFGLTGCTKK1240 pKa = 9.47LTLVSGLDD1248 pKa = 3.58YY1249 pKa = 8.76PTASSYY1255 pKa = 11.69SLTITAVDD1263 pKa = 3.2SGATPRR1269 pKa = 11.84TGSVVVTVTVTDD1281 pKa = 3.6VNRR1284 pKa = 11.84YY1285 pKa = 6.85TPSCSPATYY1294 pKa = 9.74SASVLEE1300 pKa = 4.56TAASFPTALTPATTITCTDD1319 pKa = 3.05GDD1321 pKa = 4.52GNGVYY1326 pKa = 9.89YY1327 pKa = 10.26QLVDD1331 pKa = 3.39SSVPFSVNNLTGVLSLNGPVDD1352 pKa = 3.67YY1353 pKa = 11.04DD1354 pKa = 3.84SAVQTYY1360 pKa = 8.29TILIRR1365 pKa = 11.84AVDD1368 pKa = 3.84PADD1371 pKa = 3.58SSKK1374 pKa = 11.33SGLATATVAVTAANEE1389 pKa = 4.04FTPTFPTATVTLSVYY1404 pKa = 8.32EE1405 pKa = 4.29TVASSTTITTFAATDD1420 pKa = 3.45ADD1422 pKa = 4.25YY1423 pKa = 11.13SPHH1426 pKa = 7.41AITSYY1431 pKa = 11.17AIASGNTGGMFAISNAGVITVASGMTIDD1459 pKa = 4.03YY1460 pKa = 8.33EE1461 pKa = 4.38AVSGSSPYY1469 pKa = 10.56VLQIVATDD1477 pKa = 3.6GGGLTGTGTISIVILDD1493 pKa = 4.4SNDD1496 pKa = 3.41LPPVFSPANYY1506 pKa = 9.14SASVPEE1512 pKa = 4.64GSAAGTTVTTVTATDD1527 pKa = 3.81GDD1529 pKa = 4.04TALNNNNKK1537 pKa = 9.27ILYY1540 pKa = 9.95SKK1542 pKa = 9.93LTGDD1546 pKa = 3.28SAGYY1550 pKa = 9.96FSVNSTNGLILTTGNILNYY1569 pKa = 9.34DD1570 pKa = 3.69TVTQVVFTVTAVDD1583 pKa = 3.82GNGAGPHH1590 pKa = 5.67TATTTVTVAVTDD1602 pKa = 4.2VNLHH1606 pKa = 6.5APSCSPSTVTVNNIRR1621 pKa = 11.84EE1622 pKa = 4.22DD1623 pKa = 3.87VTVGTALTSLTSPVVICTDD1642 pKa = 3.34GDD1644 pKa = 3.84AKK1646 pKa = 10.62DD1647 pKa = 3.48TLTYY1651 pKa = 10.42TMSASASDD1659 pKa = 3.02SHH1661 pKa = 6.92FAVNPTSGAITVSTALDD1678 pKa = 3.55FEE1680 pKa = 4.84TARR1683 pKa = 11.84IHH1685 pKa = 5.09QFEE1688 pKa = 4.54VVVTDD1693 pKa = 3.26SATPAKK1699 pKa = 9.27SVSMAVTINVLDD1711 pKa = 3.9VNEE1714 pKa = 4.81FSPTCTVPTTASMNEE1729 pKa = 3.73AQAVGTAAYY1738 pKa = 9.91SATSCSDD1745 pKa = 3.07SDD1747 pKa = 3.89GTATTLQFALVPGTPFQVSSSGAVTASSALDD1778 pKa = 3.73YY1779 pKa = 11.17DD1780 pKa = 4.16PASATTSYY1788 pKa = 10.77SVRR1791 pKa = 11.84LIVSDD1796 pKa = 3.82NAPTISSLRR1805 pKa = 11.84SSTYY1809 pKa = 6.93TTVVSVTPVNEE1820 pKa = 4.02FAPVLTYY1827 pKa = 9.55TALTASLAEE1836 pKa = 4.35DD1837 pKa = 3.82SGTGTALALKK1847 pKa = 8.8TIGGTSVSMAATDD1860 pKa = 3.67ADD1862 pKa = 3.96RR1863 pKa = 11.84GLNHH1867 pKa = 7.11ATLTWSIVGGNDD1879 pKa = 3.04AGKK1882 pKa = 10.48FSINSASGALQTAAALDD1899 pKa = 3.96RR1900 pKa = 11.84EE1901 pKa = 4.64TAASYY1906 pKa = 9.73TLLVSVTDD1914 pKa = 3.93GGGLSSTANCTVAVSVTDD1932 pKa = 3.68INDD1935 pKa = 3.32NTPVCNPASQTITLYY1950 pKa = 10.56EE1951 pKa = 4.23GEE1953 pKa = 4.19AAKK1956 pKa = 10.64QLVDD1960 pKa = 4.63FGASGRR1966 pKa = 11.84VTDD1969 pKa = 4.87ADD1971 pKa = 3.75SGVNAQLTFSLNNAAALSPDD1991 pKa = 3.46FQLSGTRR1998 pKa = 11.84LSSVRR2003 pKa = 11.84SFSSSDD2009 pKa = 3.33TLSNTLLVVATDD2021 pKa = 4.2GGSSARR2027 pKa = 11.84SATCSVTVNIAVTARR2042 pKa = 11.84PPTFTVANSAVTVSEE2057 pKa = 4.26SSSVGTQVFQFVPNLGTGGTGPLTMSLVDD2086 pKa = 4.94FSPSANSNLFAFDD2099 pKa = 3.58ASTGRR2104 pKa = 11.84VSVNQALDD2112 pKa = 3.65YY2113 pKa = 10.81DD2114 pKa = 4.36SSSTRR2119 pKa = 11.84VPLSS2123 pKa = 3.13
MM1 pKa = 7.62APNGVSDD8 pKa = 4.2RR9 pKa = 11.84SARR12 pKa = 11.84LPRR15 pKa = 11.84LIMIRR20 pKa = 11.84ILLLVAMQATLFPGTAFAQLAWDD43 pKa = 4.14TFSPAQQTYY52 pKa = 7.7PTATVTIDD60 pKa = 3.04EE61 pKa = 4.44NMAATTTVLTITTTGATGTVTYY83 pKa = 10.26SIASSTTAAFTIGASTGVITVNDD106 pKa = 3.66ASLLDD111 pKa = 3.71RR112 pKa = 11.84EE113 pKa = 4.88AISFIDD119 pKa = 4.29LVLQAVDD126 pKa = 3.62TSPATVSTTVRR137 pKa = 11.84IDD139 pKa = 3.46LKK141 pKa = 10.77DD142 pKa = 3.65VNEE145 pKa = 4.08IAPTFSSTSTYY156 pKa = 10.81SVTISEE162 pKa = 4.55SATVGTTIQALTVTDD177 pKa = 3.69TDD179 pKa = 3.87SAATNVACTISSGDD193 pKa = 3.72PDD195 pKa = 3.64GKK197 pKa = 10.71FQFRR201 pKa = 11.84SDD203 pKa = 3.34NQTIEE208 pKa = 4.14LKK210 pKa = 10.85AAIEE214 pKa = 4.18PDD216 pKa = 3.55SPTNNSLTYY225 pKa = 9.79TLTIVCYY232 pKa = 10.79DD233 pKa = 3.69NFNGVYY239 pKa = 10.14SGGSLASTGTVSVSVTPVNDD259 pKa = 3.98NSPTWGTFTPALATPITTIDD279 pKa = 3.5EE280 pKa = 4.24NLASGTIFTAAASDD294 pKa = 3.48VDD296 pKa = 4.09YY297 pKa = 8.96GTDD300 pKa = 3.47GVVTFSFSTVKK311 pKa = 10.6FNTGADD317 pKa = 3.29ASALFTIGSSSGVVSRR333 pKa = 11.84TAATTPALDD342 pKa = 3.46RR343 pKa = 11.84EE344 pKa = 4.89TYY346 pKa = 9.25SHH348 pKa = 6.83VEE350 pKa = 4.04FVVLAMDD357 pKa = 4.43SPTVTADD364 pKa = 3.5RR365 pKa = 11.84LTITQTVQVNLKK377 pKa = 9.81DD378 pKa = 3.91VNEE381 pKa = 4.15FTPVFDD387 pKa = 5.16PSTGLYY393 pKa = 9.52NVSFSEE399 pKa = 4.21TAPAGTLVLNVSVSDD414 pKa = 3.9PDD416 pKa = 3.99FSRR419 pKa = 11.84TAISCTITSGNTLDD433 pKa = 3.9RR434 pKa = 11.84FQMRR438 pKa = 11.84ADD440 pKa = 3.71GTSLEE445 pKa = 4.92LKK447 pKa = 8.65TQIEE451 pKa = 4.42PDD453 pKa = 3.93SPTSDD458 pKa = 4.1PINYY462 pKa = 9.36VLTLSCADD470 pKa = 3.66NTAGTYY476 pKa = 10.31SGGQKK481 pKa = 10.3SSTGTVTVWVEE492 pKa = 3.66YY493 pKa = 11.02NNDD496 pKa = 3.57YY497 pKa = 11.24SPVWSTFSPAWTSATTPISTILEE520 pKa = 4.01NLAVNQTVFTASATDD535 pKa = 4.34DD536 pKa = 3.9DD537 pKa = 5.62AGDD540 pKa = 4.65DD541 pKa = 3.93GLVVYY546 pKa = 10.13SISGAVAEE554 pKa = 4.69NGSSVMEE561 pKa = 4.03AFAINGSSGAVSIANSTVLDD581 pKa = 3.81RR582 pKa = 11.84EE583 pKa = 4.82GYY585 pKa = 9.46SYY587 pKa = 12.0VDD589 pKa = 3.35VTVEE593 pKa = 4.26ARR595 pKa = 11.84DD596 pKa = 3.9SPASGTTYY604 pKa = 10.46TITQTVRR611 pKa = 11.84IGISDD616 pKa = 3.83YY617 pKa = 11.79NEE619 pKa = 3.95FPPAFTPSTGLYY631 pKa = 8.94TPAIPEE637 pKa = 4.06TTTAGTAVQTIVATDD652 pKa = 3.64PDD654 pKa = 4.28YY655 pKa = 11.85SNTAVVCAIAAGNSLNRR672 pKa = 11.84FAINTADD679 pKa = 3.58KK680 pKa = 10.83KK681 pKa = 10.75LVEE684 pKa = 4.42TASVIQPDD692 pKa = 4.8KK693 pKa = 9.98PTLDD697 pKa = 3.3PTIYY701 pKa = 10.86LLTVHH706 pKa = 6.63CADD709 pKa = 4.41MNNGTLGTPVLTSTAYY725 pKa = 8.23ITVNVTLVNDD735 pKa = 3.79YY736 pKa = 11.47DD737 pKa = 3.76PTITLNSTTMVIPEE751 pKa = 4.35DD752 pKa = 4.05TPVSSFIVSVTADD765 pKa = 3.62DD766 pKa = 4.94LDD768 pKa = 4.08YY769 pKa = 11.75GNDD772 pKa = 3.55GTFTISLSPTTPFALTGDD790 pKa = 3.83RR791 pKa = 11.84TKK793 pKa = 11.31LLLTEE798 pKa = 4.29ALDD801 pKa = 4.37RR802 pKa = 11.84EE803 pKa = 4.73TKK805 pKa = 10.12DD806 pKa = 3.04SYY808 pKa = 10.22VIEE811 pKa = 4.26MRR813 pKa = 11.84ALDD816 pKa = 3.63QGSPARR822 pKa = 11.84SSTTTLTVSITDD834 pKa = 3.62VNDD837 pKa = 3.28NTPVCSPNTYY847 pKa = 9.75NVSVTADD854 pKa = 3.18SAATLVASIPCTDD867 pKa = 4.48ADD869 pKa = 4.88LGDD872 pKa = 4.12NQLLAYY878 pKa = 10.13SFLSGDD884 pKa = 3.55TRR886 pKa = 11.84NLTLSSTTGALTTTFLTYY904 pKa = 10.88DD905 pKa = 3.13SAAPSKK911 pKa = 9.64TLVITVRR918 pKa = 11.84DD919 pKa = 3.69SATPASAQRR928 pKa = 11.84TATVTVTVHH937 pKa = 5.35VLPRR941 pKa = 11.84NLGPPVWGTFNPPWISATTAINSVSEE967 pKa = 4.06NADD970 pKa = 2.94EE971 pKa = 4.6GFAVFTAVAIDD982 pKa = 4.18YY983 pKa = 10.81DD984 pKa = 4.28SGPDD988 pKa = 3.54GQVIYY993 pKa = 10.93SLASVTGTDD1002 pKa = 3.25GTTSVDD1008 pKa = 3.27LSSVFNISSSSGQVQVKK1025 pKa = 8.89TKK1027 pKa = 10.87GILDD1031 pKa = 3.97RR1032 pKa = 11.84EE1033 pKa = 4.3ATPAYY1038 pKa = 9.14TYY1040 pKa = 11.01INFVVQARR1048 pKa = 11.84DD1049 pKa = 3.34NPTTGTPNSIIQTVRR1064 pKa = 11.84VDD1066 pKa = 3.32VTDD1069 pKa = 4.15FNEE1072 pKa = 4.44LDD1074 pKa = 3.52PTFTGDD1080 pKa = 3.48PFSATLVEE1088 pKa = 5.13DD1089 pKa = 3.87SAAGTLVKK1097 pKa = 10.7QMVVSDD1103 pKa = 4.42PDD1105 pKa = 3.46SSNTIITCQITSGNDD1120 pKa = 2.96LSLFRR1125 pKa = 11.84TNSANSTIIEE1135 pKa = 4.82LNTAIDD1141 pKa = 3.56PDD1143 pKa = 3.76RR1144 pKa = 11.84PKK1146 pKa = 10.92NHH1148 pKa = 7.07PLSYY1152 pKa = 10.88LLIVTCLDD1160 pKa = 3.34NNGAGSYY1167 pKa = 7.87RR1168 pKa = 11.84TATANLSVTITTTNDD1183 pKa = 2.93HH1184 pKa = 6.35TPVVSIPIGSSITLPEE1200 pKa = 3.84TQAIGDD1206 pKa = 3.77VADD1209 pKa = 3.98ISVVDD1214 pKa = 3.63QDD1216 pKa = 3.98YY1217 pKa = 8.57GTDD1220 pKa = 3.48GTVACSIAPTGTFGLTGCTKK1240 pKa = 9.47LTLVSGLDD1248 pKa = 3.58YY1249 pKa = 8.76PTASSYY1255 pKa = 11.69SLTITAVDD1263 pKa = 3.2SGATPRR1269 pKa = 11.84TGSVVVTVTVTDD1281 pKa = 3.6VNRR1284 pKa = 11.84YY1285 pKa = 6.85TPSCSPATYY1294 pKa = 9.74SASVLEE1300 pKa = 4.56TAASFPTALTPATTITCTDD1319 pKa = 3.05GDD1321 pKa = 4.52GNGVYY1326 pKa = 9.89YY1327 pKa = 10.26QLVDD1331 pKa = 3.39SSVPFSVNNLTGVLSLNGPVDD1352 pKa = 3.67YY1353 pKa = 11.04DD1354 pKa = 3.84SAVQTYY1360 pKa = 8.29TILIRR1365 pKa = 11.84AVDD1368 pKa = 3.84PADD1371 pKa = 3.58SSKK1374 pKa = 11.33SGLATATVAVTAANEE1389 pKa = 4.04FTPTFPTATVTLSVYY1404 pKa = 8.32EE1405 pKa = 4.29TVASSTTITTFAATDD1420 pKa = 3.45ADD1422 pKa = 4.25YY1423 pKa = 11.13SPHH1426 pKa = 7.41AITSYY1431 pKa = 11.17AIASGNTGGMFAISNAGVITVASGMTIDD1459 pKa = 4.03YY1460 pKa = 8.33EE1461 pKa = 4.38AVSGSSPYY1469 pKa = 10.56VLQIVATDD1477 pKa = 3.6GGGLTGTGTISIVILDD1493 pKa = 4.4SNDD1496 pKa = 3.41LPPVFSPANYY1506 pKa = 9.14SASVPEE1512 pKa = 4.64GSAAGTTVTTVTATDD1527 pKa = 3.81GDD1529 pKa = 4.04TALNNNNKK1537 pKa = 9.27ILYY1540 pKa = 9.95SKK1542 pKa = 9.93LTGDD1546 pKa = 3.28SAGYY1550 pKa = 9.96FSVNSTNGLILTTGNILNYY1569 pKa = 9.34DD1570 pKa = 3.69TVTQVVFTVTAVDD1583 pKa = 3.82GNGAGPHH1590 pKa = 5.67TATTTVTVAVTDD1602 pKa = 4.2VNLHH1606 pKa = 6.5APSCSPSTVTVNNIRR1621 pKa = 11.84EE1622 pKa = 4.22DD1623 pKa = 3.87VTVGTALTSLTSPVVICTDD1642 pKa = 3.34GDD1644 pKa = 3.84AKK1646 pKa = 10.62DD1647 pKa = 3.48TLTYY1651 pKa = 10.42TMSASASDD1659 pKa = 3.02SHH1661 pKa = 6.92FAVNPTSGAITVSTALDD1678 pKa = 3.55FEE1680 pKa = 4.84TARR1683 pKa = 11.84IHH1685 pKa = 5.09QFEE1688 pKa = 4.54VVVTDD1693 pKa = 3.26SATPAKK1699 pKa = 9.27SVSMAVTINVLDD1711 pKa = 3.9VNEE1714 pKa = 4.81FSPTCTVPTTASMNEE1729 pKa = 3.73AQAVGTAAYY1738 pKa = 9.91SATSCSDD1745 pKa = 3.07SDD1747 pKa = 3.89GTATTLQFALVPGTPFQVSSSGAVTASSALDD1778 pKa = 3.73YY1779 pKa = 11.17DD1780 pKa = 4.16PASATTSYY1788 pKa = 10.77SVRR1791 pKa = 11.84LIVSDD1796 pKa = 3.82NAPTISSLRR1805 pKa = 11.84SSTYY1809 pKa = 6.93TTVVSVTPVNEE1820 pKa = 4.02FAPVLTYY1827 pKa = 9.55TALTASLAEE1836 pKa = 4.35DD1837 pKa = 3.82SGTGTALALKK1847 pKa = 8.8TIGGTSVSMAATDD1860 pKa = 3.67ADD1862 pKa = 3.96RR1863 pKa = 11.84GLNHH1867 pKa = 7.11ATLTWSIVGGNDD1879 pKa = 3.04AGKK1882 pKa = 10.48FSINSASGALQTAAALDD1899 pKa = 3.96RR1900 pKa = 11.84EE1901 pKa = 4.64TAASYY1906 pKa = 9.73TLLVSVTDD1914 pKa = 3.93GGGLSSTANCTVAVSVTDD1932 pKa = 3.68INDD1935 pKa = 3.32NTPVCNPASQTITLYY1950 pKa = 10.56EE1951 pKa = 4.23GEE1953 pKa = 4.19AAKK1956 pKa = 10.64QLVDD1960 pKa = 4.63FGASGRR1966 pKa = 11.84VTDD1969 pKa = 4.87ADD1971 pKa = 3.75SGVNAQLTFSLNNAAALSPDD1991 pKa = 3.46FQLSGTRR1998 pKa = 11.84LSSVRR2003 pKa = 11.84SFSSSDD2009 pKa = 3.33TLSNTLLVVATDD2021 pKa = 4.2GGSSARR2027 pKa = 11.84SATCSVTVNIAVTARR2042 pKa = 11.84PPTFTVANSAVTVSEE2057 pKa = 4.26SSSVGTQVFQFVPNLGTGGTGPLTMSLVDD2086 pKa = 4.94FSPSANSNLFAFDD2099 pKa = 3.58ASTGRR2104 pKa = 11.84VSVNQALDD2112 pKa = 3.65YY2113 pKa = 10.81DD2114 pKa = 4.36SSSTRR2119 pKa = 11.84VPLSS2123 pKa = 3.13
Molecular weight: 218.72 kDa
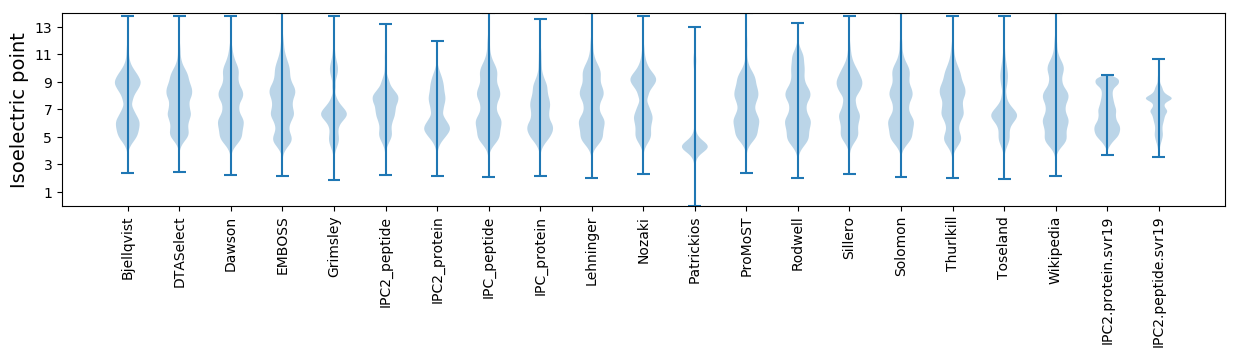
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A267DPT1|A0A267DPT1_9PLAT Uncharacterized protein OS=Macrostomum lignano OX=282301 GN=BOX15_Mlig033127g1 PE=4 SV=1
MM1 pKa = 7.24MKK3 pKa = 10.47AFSNRR8 pKa = 11.84NRR10 pKa = 11.84TNRR13 pKa = 11.84NRR15 pKa = 11.84TNRR18 pKa = 11.84NRR20 pKa = 11.84TNRR23 pKa = 11.84NRR25 pKa = 11.84TNRR28 pKa = 11.84NRR30 pKa = 11.84TNRR33 pKa = 11.84NRR35 pKa = 11.84TNRR38 pKa = 11.84NRR40 pKa = 11.84TNRR43 pKa = 11.84NRR45 pKa = 11.84TNRR48 pKa = 11.84NRR50 pKa = 11.84TNRR53 pKa = 11.84NRR55 pKa = 11.84TNRR58 pKa = 11.84KK59 pKa = 7.81RR60 pKa = 11.84KK61 pKa = 8.56NRR63 pKa = 11.84NRR65 pKa = 11.84INWNRR70 pKa = 11.84TNILIEE76 pKa = 4.28SSLLPP81 pKa = 4.17
MM1 pKa = 7.24MKK3 pKa = 10.47AFSNRR8 pKa = 11.84NRR10 pKa = 11.84TNRR13 pKa = 11.84NRR15 pKa = 11.84TNRR18 pKa = 11.84NRR20 pKa = 11.84TNRR23 pKa = 11.84NRR25 pKa = 11.84TNRR28 pKa = 11.84NRR30 pKa = 11.84TNRR33 pKa = 11.84NRR35 pKa = 11.84TNRR38 pKa = 11.84NRR40 pKa = 11.84TNRR43 pKa = 11.84NRR45 pKa = 11.84TNRR48 pKa = 11.84NRR50 pKa = 11.84TNRR53 pKa = 11.84NRR55 pKa = 11.84TNRR58 pKa = 11.84KK59 pKa = 7.81RR60 pKa = 11.84KK61 pKa = 8.56NRR63 pKa = 11.84NRR65 pKa = 11.84INWNRR70 pKa = 11.84TNILIEE76 pKa = 4.28SSLLPP81 pKa = 4.17
Molecular weight: 10.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
23504603 |
49 |
7972 |
540.8 |
59.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.943 ± 0.021 | 2.268 ± 0.009 |
5.476 ± 0.01 | 5.805 ± 0.018 |
3.443 ± 0.009 | 6.527 ± 0.015 |
2.209 ± 0.006 | 3.636 ± 0.009 |
4.204 ± 0.015 | 10.01 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.874 ± 0.005 | 3.383 ± 0.008 |
5.533 ± 0.015 | 5.3 ± 0.017 |
7.071 ± 0.019 | 8.837 ± 0.014 |
4.989 ± 0.02 | 6.005 ± 0.009 |
1.099 ± 0.004 | 2.389 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
