
Primorskyibacter insulae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pseudoprimorskyibacter
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
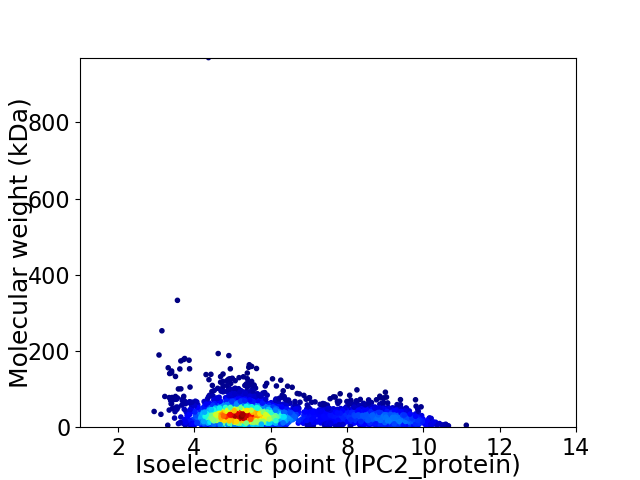
Virtual 2D-PAGE plot for 3838 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R8AWS6|A0A2R8AWS6_9RHOB Putative phosphoadenosine phosphosulfate reductase OS=Primorskyibacter insulae OX=1695997 GN=cysH PE=3 SV=1
MM1 pKa = 7.86ASPAFATFTNSLSGNYY17 pKa = 10.24DD18 pKa = 3.1EE19 pKa = 6.41DD20 pKa = 5.38RR21 pKa = 11.84YY22 pKa = 10.79ILQLTAGKK30 pKa = 10.18NYY32 pKa = 9.8WYY34 pKa = 9.96DD35 pKa = 3.37LEE37 pKa = 4.9RR38 pKa = 11.84AVDD41 pKa = 3.87DD42 pKa = 4.35PGGISVSFASQTGARR57 pKa = 11.84MDD59 pKa = 4.21GNTATADD66 pKa = 3.43GDD68 pKa = 3.68HH69 pKa = 6.79RR70 pKa = 11.84LVLKK74 pKa = 9.15SQPYY78 pKa = 10.95AMYY81 pKa = 9.31TPNYY85 pKa = 7.9VPGYY89 pKa = 9.37AGAYY93 pKa = 5.82TLSVYY98 pKa = 10.84EE99 pKa = 4.12EE100 pKa = 4.32VGNSNASTEE109 pKa = 4.06RR110 pKa = 11.84LEE112 pKa = 5.13VEE114 pKa = 4.03TAYY117 pKa = 10.37HH118 pKa = 6.37GRR120 pKa = 11.84FDD122 pKa = 3.98YY123 pKa = 11.42VSDD126 pKa = 3.75RR127 pKa = 11.84DD128 pKa = 3.86VIGFQGRR135 pKa = 11.84AGVGYY140 pKa = 9.88FNEE143 pKa = 4.09IVTEE147 pKa = 4.16VEE149 pKa = 4.02GATVSGYY156 pKa = 10.24IKK158 pKa = 10.62NDD160 pKa = 2.96GSGPSYY166 pKa = 10.86TYY168 pKa = 7.34FTPEE172 pKa = 4.07ADD174 pKa = 3.2EE175 pKa = 4.11LLYY178 pKa = 11.09VGVYY182 pKa = 9.97SGVVTDD188 pKa = 3.41YY189 pKa = 10.73TLTVRR194 pKa = 11.84QEE196 pKa = 4.06VGNSLSSVAVLKK208 pKa = 10.37PDD210 pKa = 3.2TSIASRR216 pKa = 11.84IDD218 pKa = 3.18YY219 pKa = 9.96MGDD222 pKa = 2.97YY223 pKa = 10.91DD224 pKa = 4.26VFHH227 pKa = 6.3VTPEE231 pKa = 4.16DD232 pKa = 3.89GFGLTVTVTAADD244 pKa = 3.71TDD246 pKa = 4.14PALGLDD252 pKa = 4.14LFAYY256 pKa = 9.72EE257 pKa = 4.33YY258 pKa = 10.64DD259 pKa = 3.37VSYY262 pKa = 11.14GVTDD266 pKa = 3.55TQFVLTGPTAITVGADD282 pKa = 3.43GPGGYY287 pKa = 9.4ILTAEE292 pKa = 4.21RR293 pKa = 11.84EE294 pKa = 4.37VLGTLDD300 pKa = 4.29SKK302 pKa = 12.11SMLTPDD308 pKa = 4.29APVHH312 pKa = 6.42DD313 pKa = 4.11TLQYY317 pKa = 11.2GGDD320 pKa = 3.06IDD322 pKa = 3.88VFRR325 pKa = 11.84FEE327 pKa = 4.38ITDD330 pKa = 3.63GADD333 pKa = 2.99YY334 pKa = 10.81LIRR337 pKa = 11.84FRR339 pKa = 11.84GDD341 pKa = 2.99PGGDD345 pKa = 2.78EE346 pKa = 4.15LTRR349 pKa = 11.84LRR351 pKa = 11.84FAIFDD356 pKa = 3.69EE357 pKa = 5.93DD358 pKa = 3.99GTQVPVNGGLGSEE371 pKa = 3.78WRR373 pKa = 11.84YY374 pKa = 8.18LTEE377 pKa = 4.18GLGAGTYY384 pKa = 8.27FARR387 pKa = 11.84LLTGQIYY394 pKa = 10.71DD395 pKa = 3.6GGDD398 pKa = 3.67YY399 pKa = 10.45ILTMTPEE406 pKa = 4.19EE407 pKa = 4.62AGDD410 pKa = 3.87AQTGSAIRR418 pKa = 11.84PGGQFAGNIDD428 pKa = 3.45EE429 pKa = 5.08WDD431 pKa = 3.85DD432 pKa = 3.8EE433 pKa = 4.41DD434 pKa = 3.44WVRR437 pKa = 11.84IVAQPGRR444 pKa = 11.84DD445 pKa = 3.35VTLSIDD451 pKa = 3.67MGSGAPTLSPIEE463 pKa = 4.12PEE465 pKa = 3.71EE466 pKa = 4.72GIFLSLVDD474 pKa = 3.58EE475 pKa = 4.87TGTVLAISTVTDD487 pKa = 3.36TSEE490 pKa = 4.09TLTFTPEE497 pKa = 3.55PGKK500 pKa = 9.46TYY502 pKa = 9.83FAKK505 pKa = 9.41VTPPPGSFSLFGPDD519 pKa = 2.16IFAWSEE525 pKa = 3.31GDD527 pKa = 2.93MGYY530 pKa = 11.02GYY532 pKa = 8.39TLSMTQNGVRR542 pKa = 11.84IGTALAEE549 pKa = 4.29VLSGSDD555 pKa = 3.43GPDD558 pKa = 3.28WITADD563 pKa = 4.08AGNDD567 pKa = 3.53TVNALDD573 pKa = 4.39GADD576 pKa = 3.54TVIAGPGDD584 pKa = 3.61DD585 pKa = 3.94VVNGGTTAADD595 pKa = 3.77LRR597 pKa = 11.84DD598 pKa = 3.91VIYY601 pKa = 10.72GGDD604 pKa = 3.78GDD606 pKa = 4.49DD607 pKa = 4.4SLVGGYY613 pKa = 10.89GNDD616 pKa = 3.67EE617 pKa = 4.11LRR619 pKa = 11.84GDD621 pKa = 4.01AGNDD625 pKa = 3.31TLEE628 pKa = 4.58GGFGADD634 pKa = 3.12TVLGGTGNDD643 pKa = 3.56VLTGGAWGDD652 pKa = 3.57MLFGGDD658 pKa = 5.18GDD660 pKa = 5.0DD661 pKa = 4.1FLNGGFGFDD670 pKa = 3.81RR671 pKa = 11.84LNGGSGADD679 pKa = 3.32QFFHH683 pKa = 7.11LGAAGHH689 pKa = 6.09GSDD692 pKa = 4.83WIQDD696 pKa = 3.36YY697 pKa = 10.93SAAEE701 pKa = 4.1GDD703 pKa = 3.81VLVYY707 pKa = 10.87GGTATKK713 pKa = 10.91GDD715 pKa = 4.08FLVQRR720 pKa = 11.84AATPGAGDD728 pKa = 3.55AAVKK732 pKa = 10.04EE733 pKa = 4.43VFITHH738 pKa = 5.85VPSNALLWALVDD750 pKa = 4.18GDD752 pKa = 4.02AQAAITVRR760 pKa = 11.84AGGVEE765 pKa = 4.07FDD767 pKa = 4.56LLAA770 pKa = 5.47
MM1 pKa = 7.86ASPAFATFTNSLSGNYY17 pKa = 10.24DD18 pKa = 3.1EE19 pKa = 6.41DD20 pKa = 5.38RR21 pKa = 11.84YY22 pKa = 10.79ILQLTAGKK30 pKa = 10.18NYY32 pKa = 9.8WYY34 pKa = 9.96DD35 pKa = 3.37LEE37 pKa = 4.9RR38 pKa = 11.84AVDD41 pKa = 3.87DD42 pKa = 4.35PGGISVSFASQTGARR57 pKa = 11.84MDD59 pKa = 4.21GNTATADD66 pKa = 3.43GDD68 pKa = 3.68HH69 pKa = 6.79RR70 pKa = 11.84LVLKK74 pKa = 9.15SQPYY78 pKa = 10.95AMYY81 pKa = 9.31TPNYY85 pKa = 7.9VPGYY89 pKa = 9.37AGAYY93 pKa = 5.82TLSVYY98 pKa = 10.84EE99 pKa = 4.12EE100 pKa = 4.32VGNSNASTEE109 pKa = 4.06RR110 pKa = 11.84LEE112 pKa = 5.13VEE114 pKa = 4.03TAYY117 pKa = 10.37HH118 pKa = 6.37GRR120 pKa = 11.84FDD122 pKa = 3.98YY123 pKa = 11.42VSDD126 pKa = 3.75RR127 pKa = 11.84DD128 pKa = 3.86VIGFQGRR135 pKa = 11.84AGVGYY140 pKa = 9.88FNEE143 pKa = 4.09IVTEE147 pKa = 4.16VEE149 pKa = 4.02GATVSGYY156 pKa = 10.24IKK158 pKa = 10.62NDD160 pKa = 2.96GSGPSYY166 pKa = 10.86TYY168 pKa = 7.34FTPEE172 pKa = 4.07ADD174 pKa = 3.2EE175 pKa = 4.11LLYY178 pKa = 11.09VGVYY182 pKa = 9.97SGVVTDD188 pKa = 3.41YY189 pKa = 10.73TLTVRR194 pKa = 11.84QEE196 pKa = 4.06VGNSLSSVAVLKK208 pKa = 10.37PDD210 pKa = 3.2TSIASRR216 pKa = 11.84IDD218 pKa = 3.18YY219 pKa = 9.96MGDD222 pKa = 2.97YY223 pKa = 10.91DD224 pKa = 4.26VFHH227 pKa = 6.3VTPEE231 pKa = 4.16DD232 pKa = 3.89GFGLTVTVTAADD244 pKa = 3.71TDD246 pKa = 4.14PALGLDD252 pKa = 4.14LFAYY256 pKa = 9.72EE257 pKa = 4.33YY258 pKa = 10.64DD259 pKa = 3.37VSYY262 pKa = 11.14GVTDD266 pKa = 3.55TQFVLTGPTAITVGADD282 pKa = 3.43GPGGYY287 pKa = 9.4ILTAEE292 pKa = 4.21RR293 pKa = 11.84EE294 pKa = 4.37VLGTLDD300 pKa = 4.29SKK302 pKa = 12.11SMLTPDD308 pKa = 4.29APVHH312 pKa = 6.42DD313 pKa = 4.11TLQYY317 pKa = 11.2GGDD320 pKa = 3.06IDD322 pKa = 3.88VFRR325 pKa = 11.84FEE327 pKa = 4.38ITDD330 pKa = 3.63GADD333 pKa = 2.99YY334 pKa = 10.81LIRR337 pKa = 11.84FRR339 pKa = 11.84GDD341 pKa = 2.99PGGDD345 pKa = 2.78EE346 pKa = 4.15LTRR349 pKa = 11.84LRR351 pKa = 11.84FAIFDD356 pKa = 3.69EE357 pKa = 5.93DD358 pKa = 3.99GTQVPVNGGLGSEE371 pKa = 3.78WRR373 pKa = 11.84YY374 pKa = 8.18LTEE377 pKa = 4.18GLGAGTYY384 pKa = 8.27FARR387 pKa = 11.84LLTGQIYY394 pKa = 10.71DD395 pKa = 3.6GGDD398 pKa = 3.67YY399 pKa = 10.45ILTMTPEE406 pKa = 4.19EE407 pKa = 4.62AGDD410 pKa = 3.87AQTGSAIRR418 pKa = 11.84PGGQFAGNIDD428 pKa = 3.45EE429 pKa = 5.08WDD431 pKa = 3.85DD432 pKa = 3.8EE433 pKa = 4.41DD434 pKa = 3.44WVRR437 pKa = 11.84IVAQPGRR444 pKa = 11.84DD445 pKa = 3.35VTLSIDD451 pKa = 3.67MGSGAPTLSPIEE463 pKa = 4.12PEE465 pKa = 3.71EE466 pKa = 4.72GIFLSLVDD474 pKa = 3.58EE475 pKa = 4.87TGTVLAISTVTDD487 pKa = 3.36TSEE490 pKa = 4.09TLTFTPEE497 pKa = 3.55PGKK500 pKa = 9.46TYY502 pKa = 9.83FAKK505 pKa = 9.41VTPPPGSFSLFGPDD519 pKa = 2.16IFAWSEE525 pKa = 3.31GDD527 pKa = 2.93MGYY530 pKa = 11.02GYY532 pKa = 8.39TLSMTQNGVRR542 pKa = 11.84IGTALAEE549 pKa = 4.29VLSGSDD555 pKa = 3.43GPDD558 pKa = 3.28WITADD563 pKa = 4.08AGNDD567 pKa = 3.53TVNALDD573 pKa = 4.39GADD576 pKa = 3.54TVIAGPGDD584 pKa = 3.61DD585 pKa = 3.94VVNGGTTAADD595 pKa = 3.77LRR597 pKa = 11.84DD598 pKa = 3.91VIYY601 pKa = 10.72GGDD604 pKa = 3.78GDD606 pKa = 4.49DD607 pKa = 4.4SLVGGYY613 pKa = 10.89GNDD616 pKa = 3.67EE617 pKa = 4.11LRR619 pKa = 11.84GDD621 pKa = 4.01AGNDD625 pKa = 3.31TLEE628 pKa = 4.58GGFGADD634 pKa = 3.12TVLGGTGNDD643 pKa = 3.56VLTGGAWGDD652 pKa = 3.57MLFGGDD658 pKa = 5.18GDD660 pKa = 5.0DD661 pKa = 4.1FLNGGFGFDD670 pKa = 3.81RR671 pKa = 11.84LNGGSGADD679 pKa = 3.32QFFHH683 pKa = 7.11LGAAGHH689 pKa = 6.09GSDD692 pKa = 4.83WIQDD696 pKa = 3.36YY697 pKa = 10.93SAAEE701 pKa = 4.1GDD703 pKa = 3.81VLVYY707 pKa = 10.87GGTATKK713 pKa = 10.91GDD715 pKa = 4.08FLVQRR720 pKa = 11.84AATPGAGDD728 pKa = 3.55AAVKK732 pKa = 10.04EE733 pKa = 4.43VFITHH738 pKa = 5.85VPSNALLWALVDD750 pKa = 4.18GDD752 pKa = 4.02AQAAITVRR760 pKa = 11.84AGGVEE765 pKa = 4.07FDD767 pKa = 4.56LLAA770 pKa = 5.47
Molecular weight: 80.98 kDa
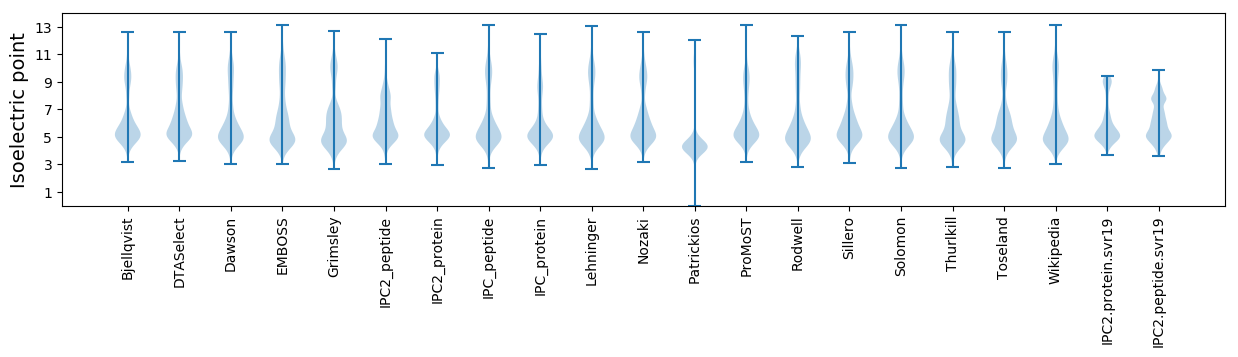
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R8APN7|A0A2R8APN7_9RHOB 4 5-dihydroxyphthalate dehydrogenase OS=Primorskyibacter insulae OX=1695997 GN=pht4 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1215343 |
29 |
9594 |
316.7 |
34.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.074 ± 0.061 | 0.869 ± 0.015 |
6.48 ± 0.05 | 5.616 ± 0.044 |
3.698 ± 0.029 | 8.81 ± 0.074 |
2.024 ± 0.022 | 5.223 ± 0.026 |
3.421 ± 0.036 | 9.83 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.83 ± 0.027 | 2.826 ± 0.03 |
4.907 ± 0.039 | 3.368 ± 0.025 |
6.28 ± 0.053 | 5.2 ± 0.036 |
5.686 ± 0.056 | 7.24 ± 0.042 |
1.345 ± 0.018 | 2.273 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
