
Pseudaminobacter manganicus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Pseudaminobacter
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
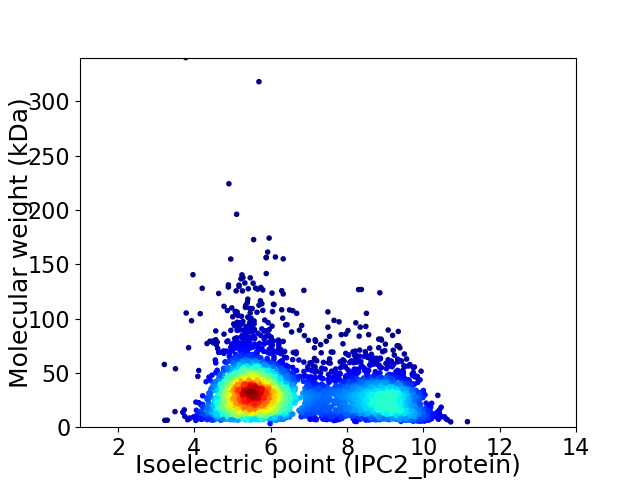
Virtual 2D-PAGE plot for 4404 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V8RQ89|A0A1V8RQ89_9RHIZ Uncharacterized protein OS=Pseudaminobacter manganicus OX=1873176 GN=BFN67_18905 PE=4 SV=1
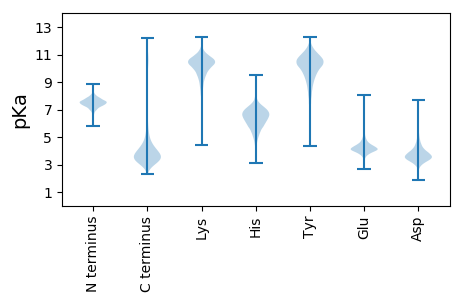
MM1 pKa = 7.65VLDD4 pKa = 4.37KK5 pKa = 11.21VSEE8 pKa = 4.73AIAHH12 pKa = 6.58FIGLFQIEE20 pKa = 4.36TEE22 pKa = 4.21QARR25 pKa = 11.84MRR27 pKa = 11.84DD28 pKa = 3.49DD29 pKa = 3.53YY30 pKa = 11.79LQFKK34 pKa = 9.91ALQAAQQPHH43 pKa = 6.6HH44 pKa = 7.42PDD46 pKa = 3.68DD47 pKa = 4.64PSQISFHH54 pKa = 6.55SPYY57 pKa = 11.03DD58 pKa = 3.68FDD60 pKa = 6.46DD61 pKa = 5.08PDD63 pKa = 5.22PGIHH67 pKa = 5.65YY68 pKa = 9.96VPNGPDD74 pKa = 3.48YY75 pKa = 11.18VSPAPVDD82 pKa = 3.19IANYY86 pKa = 7.86PQLEE90 pKa = 4.54VPVPTDD96 pKa = 3.51VQALAYY102 pKa = 8.72PGYY105 pKa = 10.08VYY107 pKa = 10.15PIIHH111 pKa = 6.91AGSHH115 pKa = 4.7SLARR119 pKa = 11.84MFEE122 pKa = 4.22LQPPGAVAAHH132 pKa = 6.77ISQVNALSDD141 pKa = 3.53NDD143 pKa = 3.88FVHH146 pKa = 6.57VGPGNVEE153 pKa = 3.74FHH155 pKa = 7.15QIGTPNLALAGLLDD169 pKa = 3.95QADD172 pKa = 3.67QALPIGPVAFGAAQDD187 pKa = 3.44IGDD190 pKa = 4.22FVSGSAAALNALVAGNEE207 pKa = 4.21GATSDD212 pKa = 5.0SIGAGDD218 pKa = 3.93GDD220 pKa = 4.18GVTIEE225 pKa = 4.75TIVSAFNEE233 pKa = 4.09PAADD237 pKa = 4.27AIYY240 pKa = 10.81VNGHH244 pKa = 5.95IVTTAPRR251 pKa = 11.84LDD253 pKa = 4.24DD254 pKa = 4.17MLPSASPLAKK264 pKa = 10.1DD265 pKa = 4.56APDD268 pKa = 4.28PAPDD272 pKa = 3.69PTPAGGNHH280 pKa = 7.63AINGGGDD287 pKa = 3.32SAAGSSAYY295 pKa = 10.13GQGALSSDD303 pKa = 3.33MSVTLGTGANALSNAASLVNDD324 pKa = 4.37ALAGGVFAVAGNHH337 pKa = 5.97ISLDD341 pKa = 3.9AIVQINAWSDD351 pKa = 3.17SDD353 pKa = 5.1SIGASLAGWNGMSHH367 pKa = 5.58QAATAAFNIAEE378 pKa = 4.42MQRR381 pKa = 11.84IDD383 pKa = 3.12TSTSDD388 pKa = 3.61GAGAAHH394 pKa = 7.2PGFPKK399 pKa = 10.24AWVVTEE405 pKa = 3.76ISGDD409 pKa = 3.84YY410 pKa = 10.98VSLNWVQQLNFVTDD424 pKa = 3.2NDD426 pKa = 4.38TAFVTSSHH434 pKa = 6.08GVTTMVGTGEE444 pKa = 4.01NQTFNNVSVADD455 pKa = 4.07LGHH458 pKa = 6.43YY459 pKa = 9.73YY460 pKa = 10.89DD461 pKa = 5.18LVLIGGNYY469 pKa = 9.29YY470 pKa = 10.37DD471 pKa = 4.93ANIIAQTNVLLDD483 pKa = 4.06DD484 pKa = 5.03DD485 pKa = 4.51VAGSIGGFQTSGTGTVSTSDD505 pKa = 3.2NLLWNDD511 pKa = 3.03ARR513 pKa = 11.84IVTVGSNTVDD523 pKa = 3.33ALPSGFAKK531 pKa = 10.65ALQDD535 pKa = 4.44FEE537 pKa = 6.76GGDD540 pKa = 3.72HH541 pKa = 6.3TLGAAVLNDD550 pKa = 3.73DD551 pKa = 5.02AFQGMAGLRR560 pKa = 11.84VLYY563 pKa = 10.05ISGSIYY569 pKa = 10.28DD570 pKa = 4.02LQYY573 pKa = 10.4IHH575 pKa = 5.79QTNVLGDD582 pKa = 3.64SDD584 pKa = 4.58QIALAMNTTQAGDD597 pKa = 3.47GGDD600 pKa = 3.25WSVTTGSDD608 pKa = 3.41ALINTARR615 pKa = 11.84IVDD618 pKa = 4.34ADD620 pKa = 4.18PAAKK624 pKa = 9.47TYY626 pKa = 11.09VGGDD630 pKa = 3.38HH631 pKa = 6.96YY632 pKa = 11.48SDD634 pKa = 3.96EE635 pKa = 5.16LLVQTDD641 pKa = 4.28IIRR644 pKa = 11.84TDD646 pKa = 3.16HH647 pKa = 6.96LLEE650 pKa = 4.42TRR652 pKa = 11.84DD653 pKa = 3.35GDD655 pKa = 4.01HH656 pKa = 6.81LVNEE660 pKa = 4.69AVAFLSDD667 pKa = 5.35DD668 pKa = 3.91MTHH671 pKa = 7.1HH672 pKa = 7.41DD673 pKa = 3.91AAYY676 pKa = 10.35ANPDD680 pKa = 3.7DD681 pKa = 5.68PISALQAPVDD691 pKa = 3.52IHH693 pKa = 6.51PAHH696 pKa = 7.09ADD698 pKa = 2.76IMQSVISS705 pKa = 3.93
MM1 pKa = 7.65VLDD4 pKa = 4.37KK5 pKa = 11.21VSEE8 pKa = 4.73AIAHH12 pKa = 6.58FIGLFQIEE20 pKa = 4.36TEE22 pKa = 4.21QARR25 pKa = 11.84MRR27 pKa = 11.84DD28 pKa = 3.49DD29 pKa = 3.53YY30 pKa = 11.79LQFKK34 pKa = 9.91ALQAAQQPHH43 pKa = 6.6HH44 pKa = 7.42PDD46 pKa = 3.68DD47 pKa = 4.64PSQISFHH54 pKa = 6.55SPYY57 pKa = 11.03DD58 pKa = 3.68FDD60 pKa = 6.46DD61 pKa = 5.08PDD63 pKa = 5.22PGIHH67 pKa = 5.65YY68 pKa = 9.96VPNGPDD74 pKa = 3.48YY75 pKa = 11.18VSPAPVDD82 pKa = 3.19IANYY86 pKa = 7.86PQLEE90 pKa = 4.54VPVPTDD96 pKa = 3.51VQALAYY102 pKa = 8.72PGYY105 pKa = 10.08VYY107 pKa = 10.15PIIHH111 pKa = 6.91AGSHH115 pKa = 4.7SLARR119 pKa = 11.84MFEE122 pKa = 4.22LQPPGAVAAHH132 pKa = 6.77ISQVNALSDD141 pKa = 3.53NDD143 pKa = 3.88FVHH146 pKa = 6.57VGPGNVEE153 pKa = 3.74FHH155 pKa = 7.15QIGTPNLALAGLLDD169 pKa = 3.95QADD172 pKa = 3.67QALPIGPVAFGAAQDD187 pKa = 3.44IGDD190 pKa = 4.22FVSGSAAALNALVAGNEE207 pKa = 4.21GATSDD212 pKa = 5.0SIGAGDD218 pKa = 3.93GDD220 pKa = 4.18GVTIEE225 pKa = 4.75TIVSAFNEE233 pKa = 4.09PAADD237 pKa = 4.27AIYY240 pKa = 10.81VNGHH244 pKa = 5.95IVTTAPRR251 pKa = 11.84LDD253 pKa = 4.24DD254 pKa = 4.17MLPSASPLAKK264 pKa = 10.1DD265 pKa = 4.56APDD268 pKa = 4.28PAPDD272 pKa = 3.69PTPAGGNHH280 pKa = 7.63AINGGGDD287 pKa = 3.32SAAGSSAYY295 pKa = 10.13GQGALSSDD303 pKa = 3.33MSVTLGTGANALSNAASLVNDD324 pKa = 4.37ALAGGVFAVAGNHH337 pKa = 5.97ISLDD341 pKa = 3.9AIVQINAWSDD351 pKa = 3.17SDD353 pKa = 5.1SIGASLAGWNGMSHH367 pKa = 5.58QAATAAFNIAEE378 pKa = 4.42MQRR381 pKa = 11.84IDD383 pKa = 3.12TSTSDD388 pKa = 3.61GAGAAHH394 pKa = 7.2PGFPKK399 pKa = 10.24AWVVTEE405 pKa = 3.76ISGDD409 pKa = 3.84YY410 pKa = 10.98VSLNWVQQLNFVTDD424 pKa = 3.2NDD426 pKa = 4.38TAFVTSSHH434 pKa = 6.08GVTTMVGTGEE444 pKa = 4.01NQTFNNVSVADD455 pKa = 4.07LGHH458 pKa = 6.43YY459 pKa = 9.73YY460 pKa = 10.89DD461 pKa = 5.18LVLIGGNYY469 pKa = 9.29YY470 pKa = 10.37DD471 pKa = 4.93ANIIAQTNVLLDD483 pKa = 4.06DD484 pKa = 5.03DD485 pKa = 4.51VAGSIGGFQTSGTGTVSTSDD505 pKa = 3.2NLLWNDD511 pKa = 3.03ARR513 pKa = 11.84IVTVGSNTVDD523 pKa = 3.33ALPSGFAKK531 pKa = 10.65ALQDD535 pKa = 4.44FEE537 pKa = 6.76GGDD540 pKa = 3.72HH541 pKa = 6.3TLGAAVLNDD550 pKa = 3.73DD551 pKa = 5.02AFQGMAGLRR560 pKa = 11.84VLYY563 pKa = 10.05ISGSIYY569 pKa = 10.28DD570 pKa = 4.02LQYY573 pKa = 10.4IHH575 pKa = 5.79QTNVLGDD582 pKa = 3.64SDD584 pKa = 4.58QIALAMNTTQAGDD597 pKa = 3.47GGDD600 pKa = 3.25WSVTTGSDD608 pKa = 3.41ALINTARR615 pKa = 11.84IVDD618 pKa = 4.34ADD620 pKa = 4.18PAAKK624 pKa = 9.47TYY626 pKa = 11.09VGGDD630 pKa = 3.38HH631 pKa = 6.96YY632 pKa = 11.48SDD634 pKa = 3.96EE635 pKa = 5.16LLVQTDD641 pKa = 4.28IIRR644 pKa = 11.84TDD646 pKa = 3.16HH647 pKa = 6.96LLEE650 pKa = 4.42TRR652 pKa = 11.84DD653 pKa = 3.35GDD655 pKa = 4.01HH656 pKa = 6.81LVNEE660 pKa = 4.69AVAFLSDD667 pKa = 5.35DD668 pKa = 3.91MTHH671 pKa = 7.1HH672 pKa = 7.41DD673 pKa = 3.91AAYY676 pKa = 10.35ANPDD680 pKa = 3.7DD681 pKa = 5.68PISALQAPVDD691 pKa = 3.52IHH693 pKa = 6.51PAHH696 pKa = 7.09ADD698 pKa = 2.76IMQSVISS705 pKa = 3.93
Molecular weight: 73.22 kDa
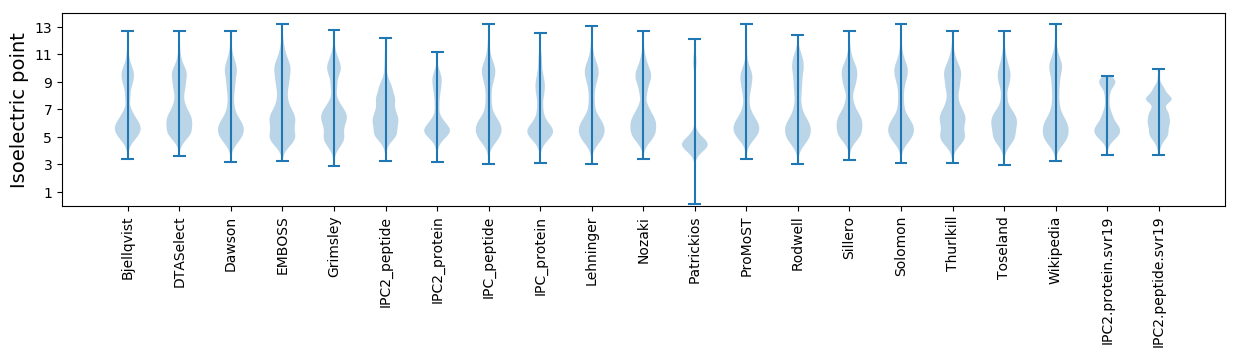
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V8RTR6|A0A1V8RTR6_9RHIZ Oxidoreductase OS=Pseudaminobacter manganicus OX=1873176 GN=BFN67_14985 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.9GGRR28 pKa = 11.84GVIVARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.9GGRR28 pKa = 11.84GVIVARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1361994 |
29 |
3478 |
309.3 |
33.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.326 ± 0.046 | 0.8 ± 0.011 |
5.735 ± 0.028 | 5.749 ± 0.035 |
3.843 ± 0.025 | 8.634 ± 0.051 |
2.072 ± 0.019 | 5.457 ± 0.023 |
3.692 ± 0.031 | 9.919 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.578 ± 0.017 | 2.689 ± 0.021 |
4.966 ± 0.029 | 2.989 ± 0.022 |
6.949 ± 0.04 | 5.444 ± 0.025 |
5.23 ± 0.029 | 7.36 ± 0.027 |
1.303 ± 0.014 | 2.265 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
