
Changjiang picorna-like virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
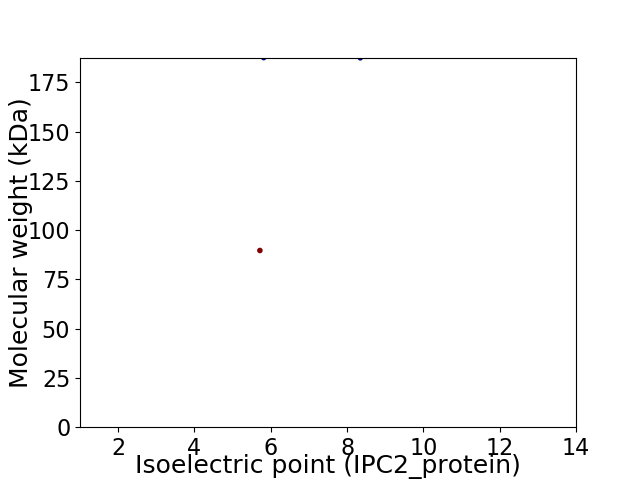
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KNP2|A0A1L3KNP2_9VIRU Uncharacterized protein OS=Changjiang picorna-like virus 10 OX=1922783 PE=4 SV=1
MM1 pKa = 7.43LRR3 pKa = 11.84DD4 pKa = 3.78DD5 pKa = 5.36TEE7 pKa = 4.3LCSNLEE13 pKa = 4.08GLHH16 pKa = 5.23MTGDD20 pKa = 3.94TKK22 pKa = 10.78TVTGVTTLLSDD33 pKa = 3.34ACEE36 pKa = 4.15STEE39 pKa = 4.39VLGKK43 pKa = 9.91HH44 pKa = 5.0YY45 pKa = 11.04APGKK49 pKa = 8.23VVRR52 pKa = 11.84SQPEE56 pKa = 3.89LQDD59 pKa = 3.19LKK61 pKa = 11.2EE62 pKa = 4.04YY63 pKa = 10.38FRR65 pKa = 11.84RR66 pKa = 11.84PRR68 pKa = 11.84IIASGALPNSVRR80 pKa = 11.84NLFNQWEE87 pKa = 4.52FTSANIFSTWFPGGITRR104 pKa = 11.84LLGVYY109 pKa = 9.23GCKK112 pKa = 10.26FKK114 pKa = 10.79IVFTLQVAATPFHH127 pKa = 6.92QGLLCLNWQYY137 pKa = 10.38GTTAGSTVTMPRR149 pKa = 11.84NKK151 pKa = 9.99FSPLSTNIPHH161 pKa = 6.03VRR163 pKa = 11.84LDD165 pKa = 3.72LAEE168 pKa = 4.11STMAQLTVPFLYY180 pKa = 10.1EE181 pKa = 3.99FEE183 pKa = 4.18FMPIGSNNTPPYY195 pKa = 10.31GILGLNLLLPIPVVVGINAPTYY217 pKa = 9.84KK218 pKa = 10.68LLVHH222 pKa = 6.81LEE224 pKa = 3.89DD225 pKa = 5.08LEE227 pKa = 5.41LIGASHH233 pKa = 7.61FATSTVVAQAGRR245 pKa = 11.84KK246 pKa = 6.74MSPIKK251 pKa = 10.71EE252 pKa = 4.12EE253 pKa = 4.43FEE255 pKa = 3.92NDD257 pKa = 3.02AYY259 pKa = 10.13PYY261 pKa = 10.98SSTLHH266 pKa = 5.62SASRR270 pKa = 11.84TLGWIAKK277 pKa = 7.76GIPSLASLTAPAAWFLDD294 pKa = 3.26KK295 pKa = 11.11SAGVLRR301 pKa = 11.84SYY303 pKa = 11.4GFAKK307 pKa = 10.4PLVQDD312 pKa = 3.65PPMRR316 pKa = 11.84AQKK319 pKa = 10.84LLTANEE325 pKa = 4.03QVVDD329 pKa = 4.23VPSTALMVAPFASNSLSVDD348 pKa = 3.14AQFGGTAVDD357 pKa = 3.71EE358 pKa = 4.29MSLAFINSRR367 pKa = 11.84WAQICVGSISTARR380 pKa = 11.84NTGTVIYY387 pKa = 8.54ATGVGPSYY395 pKa = 10.55FWFRR399 pKa = 11.84TRR401 pKa = 11.84ANPPFCNVPVPAYY414 pKa = 10.44SATGNNGILPSHH426 pKa = 6.92LLFTSSCFKK435 pKa = 10.5LWRR438 pKa = 11.84GGFKK442 pKa = 10.58FRR444 pKa = 11.84FTFSKK449 pKa = 9.84TKK451 pKa = 10.08LHH453 pKa = 6.34GGRR456 pKa = 11.84MLVAYY461 pKa = 9.0FPYY464 pKa = 10.9SNLGSNTAITGTTSAPVLPTYY485 pKa = 9.75LAPEE489 pKa = 4.62FASSAPQPTGLSTIFDD505 pKa = 4.26LRR507 pKa = 11.84DD508 pKa = 3.1SSIFEE513 pKa = 4.06FEE515 pKa = 4.34VPYY518 pKa = 10.64SSYY521 pKa = 10.24VPHH524 pKa = 6.19MRR526 pKa = 11.84FNEE529 pKa = 4.06LMGSLTMSVLDD540 pKa = 3.74NVQATGVVSDD550 pKa = 4.92TIDD553 pKa = 3.76FMVEE557 pKa = 3.75VCGGPDD563 pKa = 3.4FEE565 pKa = 5.51LSVPIGPIFAASQAGTPVAQSGRR588 pKa = 11.84VLTSTATDD596 pKa = 3.35VCEE599 pKa = 4.04LTIGEE604 pKa = 4.91CVTSVKK610 pKa = 10.12QLIMIPKK617 pKa = 7.92WRR619 pKa = 11.84SDD621 pKa = 3.14SAYY624 pKa = 10.56AGATYY629 pKa = 10.26GGNVYY634 pKa = 9.3PWWYY638 pKa = 10.93SPVQSVLTPGPTSFPSEE655 pKa = 4.1TFGFGAYY662 pKa = 9.87FSSAYY667 pKa = 10.37NYY669 pKa = 10.73ARR671 pKa = 11.84GSTEE675 pKa = 3.39VHH677 pKa = 6.56AYY679 pKa = 9.91AINAANSAGVVWIASMNPANNGAVPTDD706 pKa = 3.4NSPNTGTGSNAPRR719 pKa = 11.84VIATDD724 pKa = 3.34NSALHH729 pKa = 5.97VRR731 pKa = 11.84FPSYY735 pKa = 11.11QNIVRR740 pKa = 11.84YY741 pKa = 8.41GANDD745 pKa = 3.17ITSDD749 pKa = 3.09WSFEE753 pKa = 4.1LDD755 pKa = 3.18NVAKK759 pKa = 10.13IQAPLTVPNNNLIGPAVGMITMQNIGVTTITYY791 pKa = 9.41ISRR794 pKa = 11.84AAGEE798 pKa = 4.58DD799 pKa = 3.39GMLAHH804 pKa = 6.61YY805 pKa = 9.28LGPPTFILPGVVAASGGYY823 pKa = 10.06DD824 pKa = 3.56PDD826 pKa = 3.78MVGLKK831 pKa = 10.04PYY833 pKa = 10.79
MM1 pKa = 7.43LRR3 pKa = 11.84DD4 pKa = 3.78DD5 pKa = 5.36TEE7 pKa = 4.3LCSNLEE13 pKa = 4.08GLHH16 pKa = 5.23MTGDD20 pKa = 3.94TKK22 pKa = 10.78TVTGVTTLLSDD33 pKa = 3.34ACEE36 pKa = 4.15STEE39 pKa = 4.39VLGKK43 pKa = 9.91HH44 pKa = 5.0YY45 pKa = 11.04APGKK49 pKa = 8.23VVRR52 pKa = 11.84SQPEE56 pKa = 3.89LQDD59 pKa = 3.19LKK61 pKa = 11.2EE62 pKa = 4.04YY63 pKa = 10.38FRR65 pKa = 11.84RR66 pKa = 11.84PRR68 pKa = 11.84IIASGALPNSVRR80 pKa = 11.84NLFNQWEE87 pKa = 4.52FTSANIFSTWFPGGITRR104 pKa = 11.84LLGVYY109 pKa = 9.23GCKK112 pKa = 10.26FKK114 pKa = 10.79IVFTLQVAATPFHH127 pKa = 6.92QGLLCLNWQYY137 pKa = 10.38GTTAGSTVTMPRR149 pKa = 11.84NKK151 pKa = 9.99FSPLSTNIPHH161 pKa = 6.03VRR163 pKa = 11.84LDD165 pKa = 3.72LAEE168 pKa = 4.11STMAQLTVPFLYY180 pKa = 10.1EE181 pKa = 3.99FEE183 pKa = 4.18FMPIGSNNTPPYY195 pKa = 10.31GILGLNLLLPIPVVVGINAPTYY217 pKa = 9.84KK218 pKa = 10.68LLVHH222 pKa = 6.81LEE224 pKa = 3.89DD225 pKa = 5.08LEE227 pKa = 5.41LIGASHH233 pKa = 7.61FATSTVVAQAGRR245 pKa = 11.84KK246 pKa = 6.74MSPIKK251 pKa = 10.71EE252 pKa = 4.12EE253 pKa = 4.43FEE255 pKa = 3.92NDD257 pKa = 3.02AYY259 pKa = 10.13PYY261 pKa = 10.98SSTLHH266 pKa = 5.62SASRR270 pKa = 11.84TLGWIAKK277 pKa = 7.76GIPSLASLTAPAAWFLDD294 pKa = 3.26KK295 pKa = 11.11SAGVLRR301 pKa = 11.84SYY303 pKa = 11.4GFAKK307 pKa = 10.4PLVQDD312 pKa = 3.65PPMRR316 pKa = 11.84AQKK319 pKa = 10.84LLTANEE325 pKa = 4.03QVVDD329 pKa = 4.23VPSTALMVAPFASNSLSVDD348 pKa = 3.14AQFGGTAVDD357 pKa = 3.71EE358 pKa = 4.29MSLAFINSRR367 pKa = 11.84WAQICVGSISTARR380 pKa = 11.84NTGTVIYY387 pKa = 8.54ATGVGPSYY395 pKa = 10.55FWFRR399 pKa = 11.84TRR401 pKa = 11.84ANPPFCNVPVPAYY414 pKa = 10.44SATGNNGILPSHH426 pKa = 6.92LLFTSSCFKK435 pKa = 10.5LWRR438 pKa = 11.84GGFKK442 pKa = 10.58FRR444 pKa = 11.84FTFSKK449 pKa = 9.84TKK451 pKa = 10.08LHH453 pKa = 6.34GGRR456 pKa = 11.84MLVAYY461 pKa = 9.0FPYY464 pKa = 10.9SNLGSNTAITGTTSAPVLPTYY485 pKa = 9.75LAPEE489 pKa = 4.62FASSAPQPTGLSTIFDD505 pKa = 4.26LRR507 pKa = 11.84DD508 pKa = 3.1SSIFEE513 pKa = 4.06FEE515 pKa = 4.34VPYY518 pKa = 10.64SSYY521 pKa = 10.24VPHH524 pKa = 6.19MRR526 pKa = 11.84FNEE529 pKa = 4.06LMGSLTMSVLDD540 pKa = 3.74NVQATGVVSDD550 pKa = 4.92TIDD553 pKa = 3.76FMVEE557 pKa = 3.75VCGGPDD563 pKa = 3.4FEE565 pKa = 5.51LSVPIGPIFAASQAGTPVAQSGRR588 pKa = 11.84VLTSTATDD596 pKa = 3.35VCEE599 pKa = 4.04LTIGEE604 pKa = 4.91CVTSVKK610 pKa = 10.12QLIMIPKK617 pKa = 7.92WRR619 pKa = 11.84SDD621 pKa = 3.14SAYY624 pKa = 10.56AGATYY629 pKa = 10.26GGNVYY634 pKa = 9.3PWWYY638 pKa = 10.93SPVQSVLTPGPTSFPSEE655 pKa = 4.1TFGFGAYY662 pKa = 9.87FSSAYY667 pKa = 10.37NYY669 pKa = 10.73ARR671 pKa = 11.84GSTEE675 pKa = 3.39VHH677 pKa = 6.56AYY679 pKa = 9.91AINAANSAGVVWIASMNPANNGAVPTDD706 pKa = 3.4NSPNTGTGSNAPRR719 pKa = 11.84VIATDD724 pKa = 3.34NSALHH729 pKa = 5.97VRR731 pKa = 11.84FPSYY735 pKa = 11.11QNIVRR740 pKa = 11.84YY741 pKa = 8.41GANDD745 pKa = 3.17ITSDD749 pKa = 3.09WSFEE753 pKa = 4.1LDD755 pKa = 3.18NVAKK759 pKa = 10.13IQAPLTVPNNNLIGPAVGMITMQNIGVTTITYY791 pKa = 9.41ISRR794 pKa = 11.84AAGEE798 pKa = 4.58DD799 pKa = 3.39GMLAHH804 pKa = 6.61YY805 pKa = 9.28LGPPTFILPGVVAASGGYY823 pKa = 10.06DD824 pKa = 3.56PDD826 pKa = 3.78MVGLKK831 pKa = 10.04PYY833 pKa = 10.79
Molecular weight: 89.65 kDa
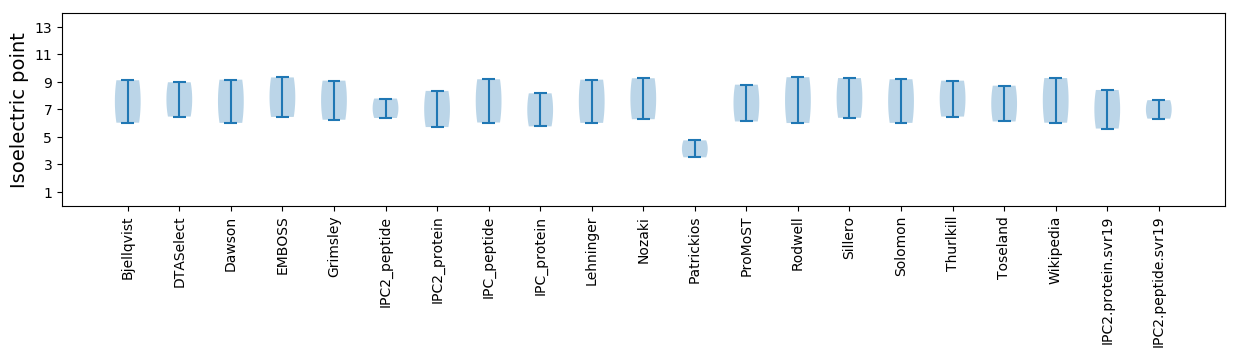
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KNP2|A0A1L3KNP2_9VIRU Uncharacterized protein OS=Changjiang picorna-like virus 10 OX=1922783 PE=4 SV=1
MM1 pKa = 7.01QAIDD5 pKa = 4.1SGSFDD10 pKa = 4.47DD11 pKa = 5.15VNIALGMRR19 pKa = 11.84VAKK22 pKa = 9.29VTFIMSMMEE31 pKa = 4.04RR32 pKa = 11.84IKK34 pKa = 10.36IDD36 pKa = 3.46EE37 pKa = 4.55ADD39 pKa = 3.87DD40 pKa = 3.85LQVRR44 pKa = 11.84HH45 pKa = 6.41LNLILDD51 pKa = 4.12AKK53 pKa = 10.34KK54 pKa = 10.53RR55 pKa = 11.84ADD57 pKa = 3.54DD58 pKa = 4.17AQDD61 pKa = 3.17EE62 pKa = 4.46RR63 pKa = 11.84EE64 pKa = 4.02RR65 pKa = 11.84AIKK68 pKa = 10.79AMDD71 pKa = 3.72ASIASMRR78 pKa = 11.84ALPQVVHH85 pKa = 6.75DD86 pKa = 4.07AWEE89 pKa = 4.33LPAMVPRR96 pKa = 11.84IRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84IKK103 pKa = 10.68VILEE107 pKa = 4.1DD108 pKa = 4.62DD109 pKa = 4.0VPKK112 pKa = 10.44PSYY115 pKa = 10.1KK116 pKa = 9.79EE117 pKa = 3.46RR118 pKa = 11.84RR119 pKa = 11.84AALVLKK125 pKa = 10.08RR126 pKa = 11.84ASRR129 pKa = 11.84TRR131 pKa = 11.84GRR133 pKa = 11.84LRR135 pKa = 11.84SALSRR140 pKa = 11.84IAVLKK145 pKa = 10.17RR146 pKa = 11.84KK147 pKa = 10.08LMDD150 pKa = 3.38RR151 pKa = 11.84VRR153 pKa = 11.84EE154 pKa = 4.16GVNVRR159 pKa = 11.84RR160 pKa = 11.84AAQAAAIAAEE170 pKa = 4.18HH171 pKa = 6.66AARR174 pKa = 11.84CAMASAKK181 pKa = 10.4GAFWTLANYY190 pKa = 10.1AGRR193 pKa = 11.84FTRR196 pKa = 11.84SMMHH200 pKa = 6.11ATSQLRR206 pKa = 11.84EE207 pKa = 4.01TMPQSGSAARR217 pKa = 11.84EE218 pKa = 3.64ARR220 pKa = 11.84ARR222 pKa = 11.84RR223 pKa = 11.84DD224 pKa = 3.45DD225 pKa = 3.49QQRR228 pKa = 11.84ASASAKK234 pKa = 9.3QRR236 pKa = 11.84AVYY239 pKa = 9.6KK240 pKa = 9.85AQPKK244 pKa = 8.57IQRR247 pKa = 11.84EE248 pKa = 3.96ADD250 pKa = 2.81IKK252 pKa = 10.17AQRR255 pKa = 11.84DD256 pKa = 3.44KK257 pKa = 11.23RR258 pKa = 11.84SGVIEE263 pKa = 4.3AEE265 pKa = 4.35SGRR268 pKa = 11.84VLPVLAGAVGATLLHH283 pKa = 6.21KK284 pKa = 10.29VWSVLTKK291 pKa = 8.1TEE293 pKa = 3.92RR294 pKa = 11.84VLTSAEE300 pKa = 3.99SLVRR304 pKa = 11.84AFKK307 pKa = 10.66KK308 pKa = 10.03GARR311 pKa = 11.84LVKK314 pKa = 9.27EE315 pKa = 4.15HH316 pKa = 6.09VGKK319 pKa = 10.64VLWVVPLVLLAHH331 pKa = 6.99RR332 pKa = 11.84MLASGEE338 pKa = 4.2FPSKK342 pKa = 10.16IAMAAIVGCLCKK354 pKa = 10.22AAGPALWKK362 pKa = 10.44HH363 pKa = 5.1IAEE366 pKa = 4.7FFPRR370 pKa = 11.84DD371 pKa = 3.35GVLEE375 pKa = 3.94QSGVGDD381 pKa = 3.54VSAKK385 pKa = 10.13LVAVVMTFTALRR397 pKa = 11.84GKK399 pKa = 9.9VCANTVTDD407 pKa = 3.69LMRR410 pKa = 11.84RR411 pKa = 11.84LSLMTRR417 pKa = 11.84ASSGLTEE424 pKa = 4.08FFDD427 pKa = 3.87WVGQAVEE434 pKa = 4.42CCVNGIRR441 pKa = 11.84QLFGKK446 pKa = 9.74EE447 pKa = 3.84AIQMYY452 pKa = 10.23KK453 pKa = 10.61DD454 pKa = 3.27SEE456 pKa = 4.32KK457 pKa = 10.62PLRR460 pKa = 11.84EE461 pKa = 3.49WMKK464 pKa = 10.77RR465 pKa = 11.84VDD467 pKa = 5.04DD468 pKa = 4.05VALSISICEE477 pKa = 4.25EE478 pKa = 4.13DD479 pKa = 4.65PSPALLNKK487 pKa = 9.88LVQLMKK493 pKa = 11.05DD494 pKa = 2.7GDD496 pKa = 4.07TFKK499 pKa = 11.07NVYY502 pKa = 10.26RR503 pKa = 11.84NTPVAKK509 pKa = 10.26RR510 pKa = 11.84IDD512 pKa = 3.76EE513 pKa = 4.62VIVKK517 pKa = 9.72VNSLLMPHH525 pKa = 6.73TGALNARR532 pKa = 11.84NNFRR536 pKa = 11.84FEE538 pKa = 4.0PDD540 pKa = 2.58ACLFLGEE547 pKa = 4.29PGIGKK552 pKa = 7.86TLLAVPLCATVLTRR566 pKa = 11.84AGLVPAAKK574 pKa = 9.34TFSEE578 pKa = 4.71VASEE582 pKa = 4.08IWQKK586 pKa = 9.57GTSEE590 pKa = 4.14FWNSYY595 pKa = 9.78SGQACVVMDD604 pKa = 4.77DD605 pKa = 3.99AFQARR610 pKa = 11.84TDD612 pKa = 3.54ASDD615 pKa = 3.51KK616 pKa = 9.95EE617 pKa = 4.25NEE619 pKa = 4.11YY620 pKa = 8.29MTLIRR625 pKa = 11.84MCSSWSYY632 pKa = 10.56PLNFADD638 pKa = 5.51LSSKK642 pKa = 10.81GKK644 pKa = 10.3IFFQSKK650 pKa = 9.94FIFGTTNVDD659 pKa = 3.99SIRR662 pKa = 11.84SSAGICISEE671 pKa = 4.06PAAVVRR677 pKa = 11.84RR678 pKa = 11.84INHH681 pKa = 6.79PYY683 pKa = 10.3RR684 pKa = 11.84LVLKK688 pKa = 10.2PEE690 pKa = 4.06YY691 pKa = 9.91KK692 pKa = 10.57SNGFLDD698 pKa = 3.38HH699 pKa = 6.91HH700 pKa = 6.98KK701 pKa = 11.22YY702 pKa = 9.73EE703 pKa = 5.38AEE705 pKa = 3.85LHH707 pKa = 6.02KK708 pKa = 10.62CAKK711 pKa = 9.38NKK713 pKa = 10.16GMDD716 pKa = 3.4HH717 pKa = 6.59FPWYY721 pKa = 7.51MWEE724 pKa = 4.03VRR726 pKa = 11.84RR727 pKa = 11.84HH728 pKa = 6.08DD729 pKa = 4.2FLSGITDD736 pKa = 3.6TVVRR740 pKa = 11.84PLKK743 pKa = 10.08EE744 pKa = 4.01VVVEE748 pKa = 5.28IIDD751 pKa = 3.93SLKK754 pKa = 10.9KK755 pKa = 9.87KK756 pKa = 8.7TDD758 pKa = 3.1SHH760 pKa = 6.57GATVAYY766 pKa = 10.42LKK768 pKa = 11.1GFVNSLQEE776 pKa = 4.04SDD778 pKa = 5.15QVIQDD783 pKa = 3.76DD784 pKa = 4.21TTVEE788 pKa = 3.86VDD790 pKa = 4.87SEE792 pKa = 4.34IKK794 pKa = 9.5PQSGGVGVKK803 pKa = 9.77LWTPYY808 pKa = 10.73SSVVMSAYY816 pKa = 9.49KK817 pKa = 10.36VYY819 pKa = 10.71KK820 pKa = 10.16SDD822 pKa = 3.89LLQKK826 pKa = 10.69FSSTQRR832 pKa = 11.84FSAALTVGVACALTVVGVRR851 pKa = 11.84LAIKK855 pKa = 8.61TALGLLHH862 pKa = 7.07ALLSGVAALFGGVKK876 pKa = 10.4SIVTGGNKK884 pKa = 8.52QPKK887 pKa = 7.96VQSNRR892 pKa = 11.84PITVPKK898 pKa = 10.09KK899 pKa = 10.72LKK901 pKa = 9.2NTDD904 pKa = 2.53IRR906 pKa = 11.84LQAGDD911 pKa = 3.64VNVINNVYY919 pKa = 10.45ANSYY923 pKa = 9.8KK924 pKa = 10.59LWFADD929 pKa = 3.61KK930 pKa = 10.4EE931 pKa = 4.74SNPVILGQVIFICDD945 pKa = 3.73RR946 pKa = 11.84LAVMPDD952 pKa = 3.01HH953 pKa = 6.89FNTVGLKK960 pKa = 10.6DD961 pKa = 3.68KK962 pKa = 10.6FDD964 pKa = 4.08EE965 pKa = 5.24GSIDD969 pKa = 4.52DD970 pKa = 3.97DD971 pKa = 4.06TVLHH975 pKa = 6.32FRR977 pKa = 11.84NAVTNAHH984 pKa = 6.18SFDD987 pKa = 3.35IKK989 pKa = 11.05YY990 pKa = 8.41GTFKK994 pKa = 10.44TFKK997 pKa = 9.91QDD999 pKa = 2.98RR1000 pKa = 11.84SEE1002 pKa = 4.22KK1003 pKa = 9.74KK1004 pKa = 9.78DD1005 pKa = 3.25VSFVVFNGVRR1015 pKa = 11.84AHH1017 pKa = 6.87RR1018 pKa = 11.84NISNAFLHH1026 pKa = 6.02QKK1028 pKa = 9.66EE1029 pKa = 3.96LDD1031 pKa = 3.92YY1032 pKa = 11.6LGGAAARR1039 pKa = 11.84LDD1041 pKa = 3.2ICEE1044 pKa = 3.69IDD1046 pKa = 3.36DD1047 pKa = 3.68RR1048 pKa = 11.84VRR1050 pKa = 11.84LRR1052 pKa = 11.84EE1053 pKa = 3.94DD1054 pKa = 3.63FARR1057 pKa = 11.84RR1058 pKa = 11.84VYY1060 pKa = 10.38VFPSVSMGKK1069 pKa = 8.66NLKK1072 pKa = 9.11YY1073 pKa = 10.53GKK1075 pKa = 8.54RR1076 pKa = 11.84TLEE1079 pKa = 3.76EE1080 pKa = 3.97YY1081 pKa = 9.34ATYY1084 pKa = 10.88KK1085 pKa = 10.12ATTSFGDD1092 pKa = 3.66CGAPASLVDD1101 pKa = 3.49NSSFGGRR1108 pKa = 11.84TVFGIHH1114 pKa = 5.29VAGNEE1119 pKa = 3.74MRR1121 pKa = 11.84GVGFSALVTSEE1132 pKa = 4.03MVKK1135 pKa = 10.59EE1136 pKa = 4.32AMAKK1140 pKa = 10.51NNIIVDD1146 pKa = 3.65NFMVDD1151 pKa = 2.92ISNHH1155 pKa = 4.31GVVAQSSYY1163 pKa = 11.33VLPPTHH1169 pKa = 6.86GGTFLPLVTIDD1180 pKa = 3.55KK1181 pKa = 10.63AVNTAVRR1188 pKa = 11.84TSYY1191 pKa = 10.74FPTEE1195 pKa = 4.01LYY1197 pKa = 9.34GTFGEE1202 pKa = 4.5YY1203 pKa = 10.3EE1204 pKa = 4.4YY1205 pKa = 11.19LPAPLAPVFRR1215 pKa = 11.84NGVLVDD1221 pKa = 3.59PMEE1224 pKa = 4.53NAVKK1228 pKa = 10.41NYY1230 pKa = 10.08SSPLFLEE1237 pKa = 4.94VPAEE1241 pKa = 3.95LDD1243 pKa = 3.6QAVHH1247 pKa = 5.35VAMSRR1252 pKa = 11.84LTAATKK1258 pKa = 10.13DD1259 pKa = 3.53YY1260 pKa = 11.1SRR1262 pKa = 11.84RR1263 pKa = 11.84IYY1265 pKa = 10.13TFEE1268 pKa = 3.87EE1269 pKa = 4.14AVLGIPQQKK1278 pKa = 9.19FRR1280 pKa = 11.84SIPRR1284 pKa = 11.84GTSAGFPYY1292 pKa = 10.22IFNVNDD1298 pKa = 3.81GKK1300 pKa = 11.29KK1301 pKa = 10.12SFFGNGQDD1309 pKa = 4.0YY1310 pKa = 11.44DD1311 pKa = 4.0LTTVKK1316 pKa = 10.63AKK1318 pKa = 9.15EE1319 pKa = 3.57LRR1321 pKa = 11.84KK1322 pKa = 9.65RR1323 pKa = 11.84VEE1325 pKa = 4.05FVEE1328 pKa = 5.31ACAQRR1333 pKa = 11.84GEE1335 pKa = 4.27RR1336 pKa = 11.84CAHH1339 pKa = 5.46VFVDD1343 pKa = 4.22FLKK1346 pKa = 11.1DD1347 pKa = 3.34EE1348 pKa = 4.13LRR1350 pKa = 11.84SRR1352 pKa = 11.84KK1353 pKa = 9.46KK1354 pKa = 10.87VEE1356 pKa = 4.13DD1357 pKa = 3.29VATRR1361 pKa = 11.84LISSAPLDD1369 pKa = 3.89YY1370 pKa = 10.76TILWRR1375 pKa = 11.84MYY1377 pKa = 9.81FGAFSSAMMNNHH1389 pKa = 6.15TLSGMAPGICTYY1401 pKa = 11.02VDD1403 pKa = 3.17WDD1405 pKa = 4.23LLAEE1409 pKa = 4.52LLRR1412 pKa = 11.84SKK1414 pKa = 11.04GSDD1417 pKa = 3.02VFAGDD1422 pKa = 3.83FKK1424 pKa = 11.56GFDD1427 pKa = 3.37ASEE1430 pKa = 4.01QEE1432 pKa = 4.28PVLMALLKK1440 pKa = 10.53YY1441 pKa = 9.7INKK1444 pKa = 9.19WYY1446 pKa = 10.08DD1447 pKa = 3.6DD1448 pKa = 4.16GPVNARR1454 pKa = 11.84VRR1456 pKa = 11.84EE1457 pKa = 4.09VLWLDD1462 pKa = 3.86LVHH1465 pKa = 6.48SRR1467 pKa = 11.84HH1468 pKa = 6.21IGGPQGDD1475 pKa = 3.67RR1476 pKa = 11.84RR1477 pKa = 11.84HH1478 pKa = 6.17IYY1480 pKa = 9.1QWNKK1484 pKa = 10.19SLPSGHH1490 pKa = 7.44PFTTICNSMYY1500 pKa = 11.13SLVTLVACYY1509 pKa = 10.05IRR1511 pKa = 11.84LTGDD1515 pKa = 3.12RR1516 pKa = 11.84TSFWTNVFTVTYY1528 pKa = 10.82GDD1530 pKa = 4.29DD1531 pKa = 3.4NVNNVNTWAASMFNQVTVADD1551 pKa = 4.08AMKK1554 pKa = 11.03SMFGLVYY1561 pKa = 10.61TSDD1564 pKa = 3.87KK1565 pKa = 10.91KK1566 pKa = 11.07DD1567 pKa = 3.17ADD1569 pKa = 3.95LVEE1572 pKa = 4.44TTVLDD1577 pKa = 3.46NVSFLKK1583 pKa = 10.62RR1584 pKa = 11.84AFEE1587 pKa = 4.39LGFDD1591 pKa = 3.69GWSCPLEE1598 pKa = 4.44LDD1600 pKa = 3.47SFLYY1604 pKa = 9.57CVYY1607 pKa = 9.4WCKK1610 pKa = 10.82NKK1612 pKa = 10.14RR1613 pKa = 11.84KK1614 pKa = 9.65KK1615 pKa = 10.62DD1616 pKa = 4.06DD1617 pKa = 4.11IIKK1620 pKa = 10.66DD1621 pKa = 3.65EE1622 pKa = 4.88LEE1624 pKa = 3.87IALQEE1629 pKa = 4.53LSLHH1633 pKa = 6.68DD1634 pKa = 3.89EE1635 pKa = 4.39KK1636 pKa = 11.62VWDD1639 pKa = 3.2KK1640 pKa = 9.74WAPRR1644 pKa = 11.84VYY1646 pKa = 10.49RR1647 pKa = 11.84LLEE1650 pKa = 4.22SRR1652 pKa = 11.84DD1653 pKa = 3.8VVPLCPCSYY1662 pKa = 9.71TEE1664 pKa = 3.67YY1665 pKa = 10.2RR1666 pKa = 11.84ALVRR1670 pKa = 11.84SRR1672 pKa = 11.84ADD1674 pKa = 2.42AWYY1677 pKa = 10.22
MM1 pKa = 7.01QAIDD5 pKa = 4.1SGSFDD10 pKa = 4.47DD11 pKa = 5.15VNIALGMRR19 pKa = 11.84VAKK22 pKa = 9.29VTFIMSMMEE31 pKa = 4.04RR32 pKa = 11.84IKK34 pKa = 10.36IDD36 pKa = 3.46EE37 pKa = 4.55ADD39 pKa = 3.87DD40 pKa = 3.85LQVRR44 pKa = 11.84HH45 pKa = 6.41LNLILDD51 pKa = 4.12AKK53 pKa = 10.34KK54 pKa = 10.53RR55 pKa = 11.84ADD57 pKa = 3.54DD58 pKa = 4.17AQDD61 pKa = 3.17EE62 pKa = 4.46RR63 pKa = 11.84EE64 pKa = 4.02RR65 pKa = 11.84AIKK68 pKa = 10.79AMDD71 pKa = 3.72ASIASMRR78 pKa = 11.84ALPQVVHH85 pKa = 6.75DD86 pKa = 4.07AWEE89 pKa = 4.33LPAMVPRR96 pKa = 11.84IRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84IKK103 pKa = 10.68VILEE107 pKa = 4.1DD108 pKa = 4.62DD109 pKa = 4.0VPKK112 pKa = 10.44PSYY115 pKa = 10.1KK116 pKa = 9.79EE117 pKa = 3.46RR118 pKa = 11.84RR119 pKa = 11.84AALVLKK125 pKa = 10.08RR126 pKa = 11.84ASRR129 pKa = 11.84TRR131 pKa = 11.84GRR133 pKa = 11.84LRR135 pKa = 11.84SALSRR140 pKa = 11.84IAVLKK145 pKa = 10.17RR146 pKa = 11.84KK147 pKa = 10.08LMDD150 pKa = 3.38RR151 pKa = 11.84VRR153 pKa = 11.84EE154 pKa = 4.16GVNVRR159 pKa = 11.84RR160 pKa = 11.84AAQAAAIAAEE170 pKa = 4.18HH171 pKa = 6.66AARR174 pKa = 11.84CAMASAKK181 pKa = 10.4GAFWTLANYY190 pKa = 10.1AGRR193 pKa = 11.84FTRR196 pKa = 11.84SMMHH200 pKa = 6.11ATSQLRR206 pKa = 11.84EE207 pKa = 4.01TMPQSGSAARR217 pKa = 11.84EE218 pKa = 3.64ARR220 pKa = 11.84ARR222 pKa = 11.84RR223 pKa = 11.84DD224 pKa = 3.45DD225 pKa = 3.49QQRR228 pKa = 11.84ASASAKK234 pKa = 9.3QRR236 pKa = 11.84AVYY239 pKa = 9.6KK240 pKa = 9.85AQPKK244 pKa = 8.57IQRR247 pKa = 11.84EE248 pKa = 3.96ADD250 pKa = 2.81IKK252 pKa = 10.17AQRR255 pKa = 11.84DD256 pKa = 3.44KK257 pKa = 11.23RR258 pKa = 11.84SGVIEE263 pKa = 4.3AEE265 pKa = 4.35SGRR268 pKa = 11.84VLPVLAGAVGATLLHH283 pKa = 6.21KK284 pKa = 10.29VWSVLTKK291 pKa = 8.1TEE293 pKa = 3.92RR294 pKa = 11.84VLTSAEE300 pKa = 3.99SLVRR304 pKa = 11.84AFKK307 pKa = 10.66KK308 pKa = 10.03GARR311 pKa = 11.84LVKK314 pKa = 9.27EE315 pKa = 4.15HH316 pKa = 6.09VGKK319 pKa = 10.64VLWVVPLVLLAHH331 pKa = 6.99RR332 pKa = 11.84MLASGEE338 pKa = 4.2FPSKK342 pKa = 10.16IAMAAIVGCLCKK354 pKa = 10.22AAGPALWKK362 pKa = 10.44HH363 pKa = 5.1IAEE366 pKa = 4.7FFPRR370 pKa = 11.84DD371 pKa = 3.35GVLEE375 pKa = 3.94QSGVGDD381 pKa = 3.54VSAKK385 pKa = 10.13LVAVVMTFTALRR397 pKa = 11.84GKK399 pKa = 9.9VCANTVTDD407 pKa = 3.69LMRR410 pKa = 11.84RR411 pKa = 11.84LSLMTRR417 pKa = 11.84ASSGLTEE424 pKa = 4.08FFDD427 pKa = 3.87WVGQAVEE434 pKa = 4.42CCVNGIRR441 pKa = 11.84QLFGKK446 pKa = 9.74EE447 pKa = 3.84AIQMYY452 pKa = 10.23KK453 pKa = 10.61DD454 pKa = 3.27SEE456 pKa = 4.32KK457 pKa = 10.62PLRR460 pKa = 11.84EE461 pKa = 3.49WMKK464 pKa = 10.77RR465 pKa = 11.84VDD467 pKa = 5.04DD468 pKa = 4.05VALSISICEE477 pKa = 4.25EE478 pKa = 4.13DD479 pKa = 4.65PSPALLNKK487 pKa = 9.88LVQLMKK493 pKa = 11.05DD494 pKa = 2.7GDD496 pKa = 4.07TFKK499 pKa = 11.07NVYY502 pKa = 10.26RR503 pKa = 11.84NTPVAKK509 pKa = 10.26RR510 pKa = 11.84IDD512 pKa = 3.76EE513 pKa = 4.62VIVKK517 pKa = 9.72VNSLLMPHH525 pKa = 6.73TGALNARR532 pKa = 11.84NNFRR536 pKa = 11.84FEE538 pKa = 4.0PDD540 pKa = 2.58ACLFLGEE547 pKa = 4.29PGIGKK552 pKa = 7.86TLLAVPLCATVLTRR566 pKa = 11.84AGLVPAAKK574 pKa = 9.34TFSEE578 pKa = 4.71VASEE582 pKa = 4.08IWQKK586 pKa = 9.57GTSEE590 pKa = 4.14FWNSYY595 pKa = 9.78SGQACVVMDD604 pKa = 4.77DD605 pKa = 3.99AFQARR610 pKa = 11.84TDD612 pKa = 3.54ASDD615 pKa = 3.51KK616 pKa = 9.95EE617 pKa = 4.25NEE619 pKa = 4.11YY620 pKa = 8.29MTLIRR625 pKa = 11.84MCSSWSYY632 pKa = 10.56PLNFADD638 pKa = 5.51LSSKK642 pKa = 10.81GKK644 pKa = 10.3IFFQSKK650 pKa = 9.94FIFGTTNVDD659 pKa = 3.99SIRR662 pKa = 11.84SSAGICISEE671 pKa = 4.06PAAVVRR677 pKa = 11.84RR678 pKa = 11.84INHH681 pKa = 6.79PYY683 pKa = 10.3RR684 pKa = 11.84LVLKK688 pKa = 10.2PEE690 pKa = 4.06YY691 pKa = 9.91KK692 pKa = 10.57SNGFLDD698 pKa = 3.38HH699 pKa = 6.91HH700 pKa = 6.98KK701 pKa = 11.22YY702 pKa = 9.73EE703 pKa = 5.38AEE705 pKa = 3.85LHH707 pKa = 6.02KK708 pKa = 10.62CAKK711 pKa = 9.38NKK713 pKa = 10.16GMDD716 pKa = 3.4HH717 pKa = 6.59FPWYY721 pKa = 7.51MWEE724 pKa = 4.03VRR726 pKa = 11.84RR727 pKa = 11.84HH728 pKa = 6.08DD729 pKa = 4.2FLSGITDD736 pKa = 3.6TVVRR740 pKa = 11.84PLKK743 pKa = 10.08EE744 pKa = 4.01VVVEE748 pKa = 5.28IIDD751 pKa = 3.93SLKK754 pKa = 10.9KK755 pKa = 9.87KK756 pKa = 8.7TDD758 pKa = 3.1SHH760 pKa = 6.57GATVAYY766 pKa = 10.42LKK768 pKa = 11.1GFVNSLQEE776 pKa = 4.04SDD778 pKa = 5.15QVIQDD783 pKa = 3.76DD784 pKa = 4.21TTVEE788 pKa = 3.86VDD790 pKa = 4.87SEE792 pKa = 4.34IKK794 pKa = 9.5PQSGGVGVKK803 pKa = 9.77LWTPYY808 pKa = 10.73SSVVMSAYY816 pKa = 9.49KK817 pKa = 10.36VYY819 pKa = 10.71KK820 pKa = 10.16SDD822 pKa = 3.89LLQKK826 pKa = 10.69FSSTQRR832 pKa = 11.84FSAALTVGVACALTVVGVRR851 pKa = 11.84LAIKK855 pKa = 8.61TALGLLHH862 pKa = 7.07ALLSGVAALFGGVKK876 pKa = 10.4SIVTGGNKK884 pKa = 8.52QPKK887 pKa = 7.96VQSNRR892 pKa = 11.84PITVPKK898 pKa = 10.09KK899 pKa = 10.72LKK901 pKa = 9.2NTDD904 pKa = 2.53IRR906 pKa = 11.84LQAGDD911 pKa = 3.64VNVINNVYY919 pKa = 10.45ANSYY923 pKa = 9.8KK924 pKa = 10.59LWFADD929 pKa = 3.61KK930 pKa = 10.4EE931 pKa = 4.74SNPVILGQVIFICDD945 pKa = 3.73RR946 pKa = 11.84LAVMPDD952 pKa = 3.01HH953 pKa = 6.89FNTVGLKK960 pKa = 10.6DD961 pKa = 3.68KK962 pKa = 10.6FDD964 pKa = 4.08EE965 pKa = 5.24GSIDD969 pKa = 4.52DD970 pKa = 3.97DD971 pKa = 4.06TVLHH975 pKa = 6.32FRR977 pKa = 11.84NAVTNAHH984 pKa = 6.18SFDD987 pKa = 3.35IKK989 pKa = 11.05YY990 pKa = 8.41GTFKK994 pKa = 10.44TFKK997 pKa = 9.91QDD999 pKa = 2.98RR1000 pKa = 11.84SEE1002 pKa = 4.22KK1003 pKa = 9.74KK1004 pKa = 9.78DD1005 pKa = 3.25VSFVVFNGVRR1015 pKa = 11.84AHH1017 pKa = 6.87RR1018 pKa = 11.84NISNAFLHH1026 pKa = 6.02QKK1028 pKa = 9.66EE1029 pKa = 3.96LDD1031 pKa = 3.92YY1032 pKa = 11.6LGGAAARR1039 pKa = 11.84LDD1041 pKa = 3.2ICEE1044 pKa = 3.69IDD1046 pKa = 3.36DD1047 pKa = 3.68RR1048 pKa = 11.84VRR1050 pKa = 11.84LRR1052 pKa = 11.84EE1053 pKa = 3.94DD1054 pKa = 3.63FARR1057 pKa = 11.84RR1058 pKa = 11.84VYY1060 pKa = 10.38VFPSVSMGKK1069 pKa = 8.66NLKK1072 pKa = 9.11YY1073 pKa = 10.53GKK1075 pKa = 8.54RR1076 pKa = 11.84TLEE1079 pKa = 3.76EE1080 pKa = 3.97YY1081 pKa = 9.34ATYY1084 pKa = 10.88KK1085 pKa = 10.12ATTSFGDD1092 pKa = 3.66CGAPASLVDD1101 pKa = 3.49NSSFGGRR1108 pKa = 11.84TVFGIHH1114 pKa = 5.29VAGNEE1119 pKa = 3.74MRR1121 pKa = 11.84GVGFSALVTSEE1132 pKa = 4.03MVKK1135 pKa = 10.59EE1136 pKa = 4.32AMAKK1140 pKa = 10.51NNIIVDD1146 pKa = 3.65NFMVDD1151 pKa = 2.92ISNHH1155 pKa = 4.31GVVAQSSYY1163 pKa = 11.33VLPPTHH1169 pKa = 6.86GGTFLPLVTIDD1180 pKa = 3.55KK1181 pKa = 10.63AVNTAVRR1188 pKa = 11.84TSYY1191 pKa = 10.74FPTEE1195 pKa = 4.01LYY1197 pKa = 9.34GTFGEE1202 pKa = 4.5YY1203 pKa = 10.3EE1204 pKa = 4.4YY1205 pKa = 11.19LPAPLAPVFRR1215 pKa = 11.84NGVLVDD1221 pKa = 3.59PMEE1224 pKa = 4.53NAVKK1228 pKa = 10.41NYY1230 pKa = 10.08SSPLFLEE1237 pKa = 4.94VPAEE1241 pKa = 3.95LDD1243 pKa = 3.6QAVHH1247 pKa = 5.35VAMSRR1252 pKa = 11.84LTAATKK1258 pKa = 10.13DD1259 pKa = 3.53YY1260 pKa = 11.1SRR1262 pKa = 11.84RR1263 pKa = 11.84IYY1265 pKa = 10.13TFEE1268 pKa = 3.87EE1269 pKa = 4.14AVLGIPQQKK1278 pKa = 9.19FRR1280 pKa = 11.84SIPRR1284 pKa = 11.84GTSAGFPYY1292 pKa = 10.22IFNVNDD1298 pKa = 3.81GKK1300 pKa = 11.29KK1301 pKa = 10.12SFFGNGQDD1309 pKa = 4.0YY1310 pKa = 11.44DD1311 pKa = 4.0LTTVKK1316 pKa = 10.63AKK1318 pKa = 9.15EE1319 pKa = 3.57LRR1321 pKa = 11.84KK1322 pKa = 9.65RR1323 pKa = 11.84VEE1325 pKa = 4.05FVEE1328 pKa = 5.31ACAQRR1333 pKa = 11.84GEE1335 pKa = 4.27RR1336 pKa = 11.84CAHH1339 pKa = 5.46VFVDD1343 pKa = 4.22FLKK1346 pKa = 11.1DD1347 pKa = 3.34EE1348 pKa = 4.13LRR1350 pKa = 11.84SRR1352 pKa = 11.84KK1353 pKa = 9.46KK1354 pKa = 10.87VEE1356 pKa = 4.13DD1357 pKa = 3.29VATRR1361 pKa = 11.84LISSAPLDD1369 pKa = 3.89YY1370 pKa = 10.76TILWRR1375 pKa = 11.84MYY1377 pKa = 9.81FGAFSSAMMNNHH1389 pKa = 6.15TLSGMAPGICTYY1401 pKa = 11.02VDD1403 pKa = 3.17WDD1405 pKa = 4.23LLAEE1409 pKa = 4.52LLRR1412 pKa = 11.84SKK1414 pKa = 11.04GSDD1417 pKa = 3.02VFAGDD1422 pKa = 3.83FKK1424 pKa = 11.56GFDD1427 pKa = 3.37ASEE1430 pKa = 4.01QEE1432 pKa = 4.28PVLMALLKK1440 pKa = 10.53YY1441 pKa = 9.7INKK1444 pKa = 9.19WYY1446 pKa = 10.08DD1447 pKa = 3.6DD1448 pKa = 4.16GPVNARR1454 pKa = 11.84VRR1456 pKa = 11.84EE1457 pKa = 4.09VLWLDD1462 pKa = 3.86LVHH1465 pKa = 6.48SRR1467 pKa = 11.84HH1468 pKa = 6.21IGGPQGDD1475 pKa = 3.67RR1476 pKa = 11.84RR1477 pKa = 11.84HH1478 pKa = 6.17IYY1480 pKa = 9.1QWNKK1484 pKa = 10.19SLPSGHH1490 pKa = 7.44PFTTICNSMYY1500 pKa = 11.13SLVTLVACYY1509 pKa = 10.05IRR1511 pKa = 11.84LTGDD1515 pKa = 3.12RR1516 pKa = 11.84TSFWTNVFTVTYY1528 pKa = 10.82GDD1530 pKa = 4.29DD1531 pKa = 3.4NVNNVNTWAASMFNQVTVADD1551 pKa = 4.08AMKK1554 pKa = 11.03SMFGLVYY1561 pKa = 10.61TSDD1564 pKa = 3.87KK1565 pKa = 10.91KK1566 pKa = 11.07DD1567 pKa = 3.17ADD1569 pKa = 3.95LVEE1572 pKa = 4.44TTVLDD1577 pKa = 3.46NVSFLKK1583 pKa = 10.62RR1584 pKa = 11.84AFEE1587 pKa = 4.39LGFDD1591 pKa = 3.69GWSCPLEE1598 pKa = 4.44LDD1600 pKa = 3.47SFLYY1604 pKa = 9.57CVYY1607 pKa = 9.4WCKK1610 pKa = 10.82NKK1612 pKa = 10.14RR1613 pKa = 11.84KK1614 pKa = 9.65KK1615 pKa = 10.62DD1616 pKa = 4.06DD1617 pKa = 4.11IIKK1620 pKa = 10.66DD1621 pKa = 3.65EE1622 pKa = 4.88LEE1624 pKa = 3.87IALQEE1629 pKa = 4.53LSLHH1633 pKa = 6.68DD1634 pKa = 3.89EE1635 pKa = 4.39KK1636 pKa = 11.62VWDD1639 pKa = 3.2KK1640 pKa = 9.74WAPRR1644 pKa = 11.84VYY1646 pKa = 10.49RR1647 pKa = 11.84LLEE1650 pKa = 4.22SRR1652 pKa = 11.84DD1653 pKa = 3.8VVPLCPCSYY1662 pKa = 9.71TEE1664 pKa = 3.67YY1665 pKa = 10.2RR1666 pKa = 11.84ALVRR1670 pKa = 11.84SRR1672 pKa = 11.84ADD1674 pKa = 2.42AWYY1677 pKa = 10.22
Molecular weight: 187.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2510 |
833 |
1677 |
1255.0 |
138.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.482 ± 0.192 | 1.474 ± 0.146 |
5.418 ± 1.036 | 4.462 ± 0.525 |
4.821 ± 0.247 | 6.614 ± 0.829 |
1.833 ± 0.146 | 4.462 ± 0.182 |
5.299 ± 1.422 | 8.606 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.59 ± 0.101 | 4.223 ± 0.502 |
4.861 ± 1.382 | 2.669 ± 0.015 |
5.697 ± 1.186 | 7.689 ± 0.703 |
6.175 ± 1.257 | 8.805 ± 0.6 |
1.554 ± 0.004 | 3.267 ± 0.307 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
