
Kushneria phosphatilytica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Kushneria
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
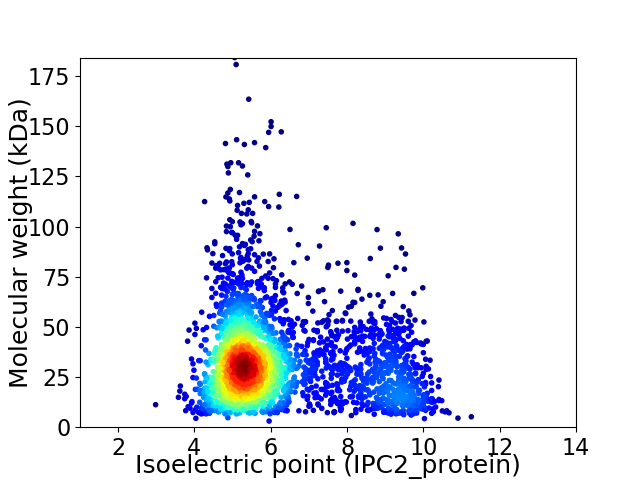
Virtual 2D-PAGE plot for 3055 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S1NXX3|A0A1S1NXX3_9GAMM Ribose-phosphate pyrophosphokinase OS=Kushneria phosphatilytica OX=657387 GN=prs PE=3 SV=1
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGVLGTLATGAQAATVYY27 pKa = 10.61NQDD30 pKa = 2.98GSKK33 pKa = 10.71LDD35 pKa = 3.46VYY37 pKa = 11.56GNVQLAFRR45 pKa = 11.84SIEE48 pKa = 4.37GYY50 pKa = 10.55DD51 pKa = 4.11DD52 pKa = 5.19SDD54 pKa = 3.75DD55 pKa = 4.08TSDD58 pKa = 4.49HH59 pKa = 5.97GTDD62 pKa = 3.32SYY64 pKa = 12.15SDD66 pKa = 3.86LFDD69 pKa = 3.98NGSTIGFRR77 pKa = 11.84GEE79 pKa = 4.66HH80 pKa = 6.77IINPGLTGYY89 pKa = 9.99FRR91 pKa = 11.84AEE93 pKa = 3.87FEE95 pKa = 4.8FNADD99 pKa = 3.51QQKK102 pKa = 10.91GYY104 pKa = 9.75YY105 pKa = 9.05AAYY108 pKa = 10.0DD109 pKa = 3.77NPDD112 pKa = 3.56DD113 pKa = 4.58QSFTEE118 pKa = 4.37SGLSRR123 pKa = 11.84GDD125 pKa = 3.32QAYY128 pKa = 10.86LGLTGNFGDD137 pKa = 4.98LRR139 pKa = 11.84IGSWDD144 pKa = 3.58SLFDD148 pKa = 3.88DD149 pKa = 5.66WIADD153 pKa = 3.99PISNNEE159 pKa = 3.88YY160 pKa = 10.66FDD162 pKa = 3.75NTDD165 pKa = 3.04AQSDD169 pKa = 4.13VGGTAYY175 pKa = 10.08EE176 pKa = 4.19SDD178 pKa = 3.87KK179 pKa = 10.45ITYY182 pKa = 7.55TSPIFSGLQFVVGTQYY198 pKa = 11.34KK199 pKa = 10.85GDD201 pKa = 3.77GEE203 pKa = 5.44DD204 pKa = 3.64YY205 pKa = 11.02NQYY208 pKa = 11.2DD209 pKa = 3.69YY210 pKa = 11.82DD211 pKa = 3.9AATGAGEE218 pKa = 5.08FVLDD222 pKa = 4.72DD223 pKa = 4.93DD224 pKa = 4.99NSASFFGGVRR234 pKa = 11.84YY235 pKa = 8.21TMAGWTLAAVYY246 pKa = 10.7DD247 pKa = 4.0DD248 pKa = 5.08LNNFEE253 pKa = 5.0YY254 pKa = 10.41ISAAGNEE261 pKa = 4.02HH262 pKa = 7.62DD263 pKa = 4.35FGKK266 pKa = 10.84LYY268 pKa = 10.66GANLSYY274 pKa = 10.72QWDD277 pKa = 4.14TLRR280 pKa = 11.84VAAKK284 pKa = 9.46WGHH287 pKa = 6.0QDD289 pKa = 4.7AKK291 pKa = 10.29WSDD294 pKa = 3.6DD295 pKa = 3.58ADD297 pKa = 3.32VDD299 pKa = 4.57RR300 pKa = 11.84YY301 pKa = 11.3GLGARR306 pKa = 11.84WGYY309 pKa = 11.51GNGDD313 pKa = 3.69LYY315 pKa = 11.5GSYY318 pKa = 10.39QYY320 pKa = 11.69VDD322 pKa = 3.69AEE324 pKa = 4.19GDD326 pKa = 3.35AAALIDD332 pKa = 3.78PRR334 pKa = 11.84RR335 pKa = 11.84FNDD338 pKa = 3.82DD339 pKa = 3.33NGNTSDD345 pKa = 3.13QHH347 pKa = 7.15YY348 pKa = 11.21NEE350 pKa = 5.58FILGATYY357 pKa = 10.91NLSSQMYY364 pKa = 9.59VFLEE368 pKa = 4.16GAKK371 pKa = 10.06YY372 pKa = 10.09DD373 pKa = 3.77RR374 pKa = 11.84KK375 pKa = 11.18DD376 pKa = 3.54NIGDD380 pKa = 3.89GVSTGMVYY388 pKa = 10.61SFF390 pKa = 4.61
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGVLGTLATGAQAATVYY27 pKa = 10.61NQDD30 pKa = 2.98GSKK33 pKa = 10.71LDD35 pKa = 3.46VYY37 pKa = 11.56GNVQLAFRR45 pKa = 11.84SIEE48 pKa = 4.37GYY50 pKa = 10.55DD51 pKa = 4.11DD52 pKa = 5.19SDD54 pKa = 3.75DD55 pKa = 4.08TSDD58 pKa = 4.49HH59 pKa = 5.97GTDD62 pKa = 3.32SYY64 pKa = 12.15SDD66 pKa = 3.86LFDD69 pKa = 3.98NGSTIGFRR77 pKa = 11.84GEE79 pKa = 4.66HH80 pKa = 6.77IINPGLTGYY89 pKa = 9.99FRR91 pKa = 11.84AEE93 pKa = 3.87FEE95 pKa = 4.8FNADD99 pKa = 3.51QQKK102 pKa = 10.91GYY104 pKa = 9.75YY105 pKa = 9.05AAYY108 pKa = 10.0DD109 pKa = 3.77NPDD112 pKa = 3.56DD113 pKa = 4.58QSFTEE118 pKa = 4.37SGLSRR123 pKa = 11.84GDD125 pKa = 3.32QAYY128 pKa = 10.86LGLTGNFGDD137 pKa = 4.98LRR139 pKa = 11.84IGSWDD144 pKa = 3.58SLFDD148 pKa = 3.88DD149 pKa = 5.66WIADD153 pKa = 3.99PISNNEE159 pKa = 3.88YY160 pKa = 10.66FDD162 pKa = 3.75NTDD165 pKa = 3.04AQSDD169 pKa = 4.13VGGTAYY175 pKa = 10.08EE176 pKa = 4.19SDD178 pKa = 3.87KK179 pKa = 10.45ITYY182 pKa = 7.55TSPIFSGLQFVVGTQYY198 pKa = 11.34KK199 pKa = 10.85GDD201 pKa = 3.77GEE203 pKa = 5.44DD204 pKa = 3.64YY205 pKa = 11.02NQYY208 pKa = 11.2DD209 pKa = 3.69YY210 pKa = 11.82DD211 pKa = 3.9AATGAGEE218 pKa = 5.08FVLDD222 pKa = 4.72DD223 pKa = 4.93DD224 pKa = 4.99NSASFFGGVRR234 pKa = 11.84YY235 pKa = 8.21TMAGWTLAAVYY246 pKa = 10.7DD247 pKa = 4.0DD248 pKa = 5.08LNNFEE253 pKa = 5.0YY254 pKa = 10.41ISAAGNEE261 pKa = 4.02HH262 pKa = 7.62DD263 pKa = 4.35FGKK266 pKa = 10.84LYY268 pKa = 10.66GANLSYY274 pKa = 10.72QWDD277 pKa = 4.14TLRR280 pKa = 11.84VAAKK284 pKa = 9.46WGHH287 pKa = 6.0QDD289 pKa = 4.7AKK291 pKa = 10.29WSDD294 pKa = 3.6DD295 pKa = 3.58ADD297 pKa = 3.32VDD299 pKa = 4.57RR300 pKa = 11.84YY301 pKa = 11.3GLGARR306 pKa = 11.84WGYY309 pKa = 11.51GNGDD313 pKa = 3.69LYY315 pKa = 11.5GSYY318 pKa = 10.39QYY320 pKa = 11.69VDD322 pKa = 3.69AEE324 pKa = 4.19GDD326 pKa = 3.35AAALIDD332 pKa = 3.78PRR334 pKa = 11.84RR335 pKa = 11.84FNDD338 pKa = 3.82DD339 pKa = 3.33NGNTSDD345 pKa = 3.13QHH347 pKa = 7.15YY348 pKa = 11.21NEE350 pKa = 5.58FILGATYY357 pKa = 10.91NLSSQMYY364 pKa = 9.59VFLEE368 pKa = 4.16GAKK371 pKa = 10.06YY372 pKa = 10.09DD373 pKa = 3.77RR374 pKa = 11.84KK375 pKa = 11.18DD376 pKa = 3.54NIGDD380 pKa = 3.89GVSTGMVYY388 pKa = 10.61SFF390 pKa = 4.61
Molecular weight: 42.87 kDa
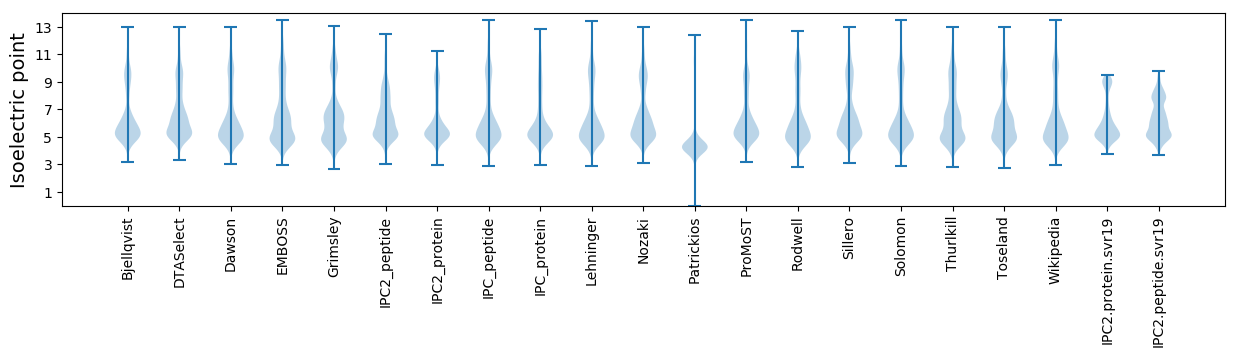
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S1NNV1|A0A1S1NNV1_9GAMM MFS domain-containing protein OS=Kushneria phosphatilytica OX=657387 GN=BH688_13075 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.5RR12 pKa = 11.84KK13 pKa = 9.44RR14 pKa = 11.84NHH16 pKa = 5.44GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.32AGRR28 pKa = 11.84QILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.5RR12 pKa = 11.84KK13 pKa = 9.44RR14 pKa = 11.84NHH16 pKa = 5.44GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.32AGRR28 pKa = 11.84QILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
948163 |
26 |
1612 |
310.4 |
34.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.503 ± 0.053 | 0.993 ± 0.014 |
5.731 ± 0.04 | 6.263 ± 0.05 |
3.418 ± 0.03 | 8.088 ± 0.041 |
2.767 ± 0.027 | 5.064 ± 0.041 |
2.239 ± 0.03 | 11.166 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.633 ± 0.022 | 2.704 ± 0.026 |
4.822 ± 0.031 | 4.18 ± 0.039 |
7.735 ± 0.046 | 5.769 ± 0.032 |
5.242 ± 0.028 | 6.741 ± 0.038 |
1.567 ± 0.021 | 2.377 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
