
Rhizobium sp. LC145
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
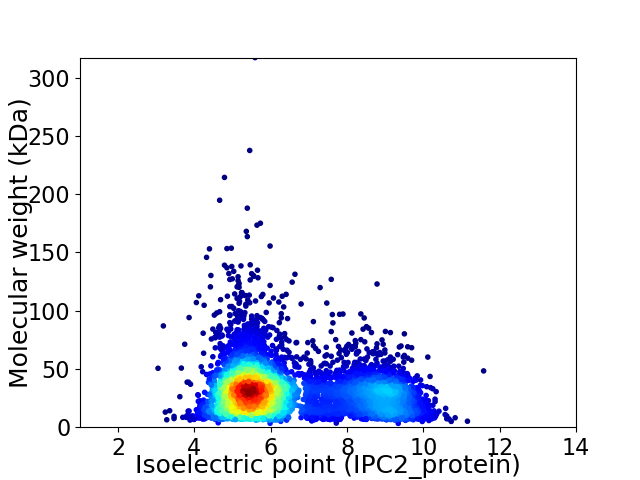
Virtual 2D-PAGE plot for 5603 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M3BKE4|A0A0M3BKE4_9RHIZ Uncharacterized protein OS=Rhizobium sp. LC145 OX=1120688 GN=YH62_11370 PE=4 SV=1
MM1 pKa = 6.78TTMDD5 pKa = 5.32KK6 pKa = 9.23ITEE9 pKa = 4.59EE10 pKa = 3.69ISHH13 pKa = 7.0FIGLFHH19 pKa = 6.01TTIEE23 pKa = 4.13DD24 pKa = 3.23VRR26 pKa = 11.84LRR28 pKa = 11.84EE29 pKa = 4.13AYY31 pKa = 9.39TDD33 pKa = 3.75FTYY36 pKa = 10.45HH37 pKa = 6.06VPQHH41 pKa = 5.94HH42 pKa = 6.67LRR44 pKa = 11.84VEE46 pKa = 4.09PVFDD50 pKa = 4.64SNFEE54 pKa = 4.04APFEE58 pKa = 4.02FLGFDD63 pKa = 3.36PGVSYY68 pKa = 10.6KK69 pKa = 10.85APAYY73 pKa = 8.86VKK75 pKa = 10.23PITHH79 pKa = 7.29PAFLSHH85 pKa = 6.55IQHH88 pKa = 6.64PPIPVVDD95 pKa = 4.92GPSEE99 pKa = 4.2YY100 pKa = 10.14PGMDD104 pKa = 3.81LPRR107 pKa = 11.84DD108 pKa = 3.93LGPAGPRR115 pKa = 11.84SMSVTLEE122 pKa = 3.52IDD124 pKa = 3.5PPGSVANYY132 pKa = 7.14MVQAAGLSDD141 pKa = 4.52DD142 pKa = 5.59DD143 pKa = 4.18YY144 pKa = 11.78FNVGGSGLAFNPEE157 pKa = 4.24PVNDD161 pKa = 3.98TDD163 pKa = 4.49LLEE166 pKa = 4.27AAEE169 pKa = 5.59DD170 pKa = 3.73ILSLSPIGDD179 pKa = 3.52PEE181 pKa = 4.26MPGSGDD187 pKa = 3.46EE188 pKa = 4.39LIEE191 pKa = 4.66LIEE194 pKa = 4.59TIASQLKK201 pKa = 9.02NWTASAEE208 pKa = 4.25SPMEE212 pKa = 4.14VFVHH216 pKa = 5.67QAATIEE222 pKa = 4.58GIYY225 pKa = 10.76VNGEE229 pKa = 4.16LVTEE233 pKa = 4.26APKK236 pKa = 10.94LSDD239 pKa = 3.16YY240 pKa = 11.31HH241 pKa = 8.58SFDD244 pKa = 4.99DD245 pKa = 5.21EE246 pKa = 4.85EE247 pKa = 5.44EE248 pKa = 5.13GDD250 pKa = 5.05DD251 pKa = 4.89DD252 pKa = 4.87TDD254 pKa = 5.24DD255 pKa = 4.41GPAPNVVVHH264 pKa = 7.07ADD266 pKa = 3.41GTVTIDD272 pKa = 3.24VSVEE276 pKa = 3.89VVAGGNTVVNNAVMQNLWTAATVTAVLGDD305 pKa = 3.7HH306 pKa = 6.69FEE308 pKa = 4.32INAIIQTNAIWDD320 pKa = 3.72TDD322 pKa = 4.34AITSAIEE329 pKa = 3.7SWTNSAGANEE339 pKa = 4.54IFNIATFEE347 pKa = 4.1RR348 pKa = 11.84MDD350 pKa = 3.47SSEE353 pKa = 4.23DD354 pKa = 3.17TGAQPVTGGFPSAWAITEE372 pKa = 3.94IQGDD376 pKa = 3.95LMIANWLEE384 pKa = 3.7QYY386 pKa = 10.76IFVSDD391 pKa = 3.69NDD393 pKa = 3.48IGILSSSGVTTSVIAGDD410 pKa = 3.72NTSTNHH416 pKa = 5.22VSLYY420 pKa = 10.55EE421 pKa = 4.22LGLSYY426 pKa = 11.29DD427 pKa = 4.49LIIVGGSVYY436 pKa = 10.41DD437 pKa = 3.82ANIIHH442 pKa = 6.0QFNLLFDD449 pKa = 4.39NDD451 pKa = 3.59VVGAVSGFEE460 pKa = 4.23TTGEE464 pKa = 4.33GAFSGAGNLLWNQAYY479 pKa = 9.54IYY481 pKa = 10.2NIGGADD487 pKa = 3.72RR488 pKa = 11.84FDD490 pKa = 4.84ALPSAYY496 pKa = 10.22LDD498 pKa = 3.7AANGIAAGRR507 pKa = 11.84NSLPNGVLTDD517 pKa = 3.74PAFGGMEE524 pKa = 4.35GLRR527 pKa = 11.84VLYY530 pKa = 10.36ISGDD534 pKa = 3.74LLNIQYY540 pKa = 9.76IRR542 pKa = 11.84QTTIVGDD549 pKa = 3.95DD550 pKa = 3.77DD551 pKa = 4.94QIALAMNAVNPHH563 pKa = 7.1PDD565 pKa = 3.24ASWSVSTGDD574 pKa = 3.25NTLVNTAAILDD585 pKa = 4.36LDD587 pKa = 4.41SMGKK591 pKa = 8.03TYY593 pKa = 11.0VGGEE597 pKa = 4.09QYY599 pKa = 10.88SQEE602 pKa = 4.08TLIQAEE608 pKa = 5.19LISSDD613 pKa = 3.88PEE615 pKa = 3.84LAARR619 pKa = 11.84DD620 pKa = 3.75PNEE623 pKa = 4.12LVNEE627 pKa = 4.1AVVFLDD633 pKa = 5.08DD634 pKa = 4.84SMLTAEE640 pKa = 4.83TDD642 pKa = 3.68AQGLPAPEE650 pKa = 5.02HH651 pKa = 6.78DD652 pKa = 3.64SHH654 pKa = 6.41YY655 pKa = 11.15QNDD658 pKa = 4.36GLQHH662 pKa = 5.34VLGG665 pKa = 4.52
MM1 pKa = 6.78TTMDD5 pKa = 5.32KK6 pKa = 9.23ITEE9 pKa = 4.59EE10 pKa = 3.69ISHH13 pKa = 7.0FIGLFHH19 pKa = 6.01TTIEE23 pKa = 4.13DD24 pKa = 3.23VRR26 pKa = 11.84LRR28 pKa = 11.84EE29 pKa = 4.13AYY31 pKa = 9.39TDD33 pKa = 3.75FTYY36 pKa = 10.45HH37 pKa = 6.06VPQHH41 pKa = 5.94HH42 pKa = 6.67LRR44 pKa = 11.84VEE46 pKa = 4.09PVFDD50 pKa = 4.64SNFEE54 pKa = 4.04APFEE58 pKa = 4.02FLGFDD63 pKa = 3.36PGVSYY68 pKa = 10.6KK69 pKa = 10.85APAYY73 pKa = 8.86VKK75 pKa = 10.23PITHH79 pKa = 7.29PAFLSHH85 pKa = 6.55IQHH88 pKa = 6.64PPIPVVDD95 pKa = 4.92GPSEE99 pKa = 4.2YY100 pKa = 10.14PGMDD104 pKa = 3.81LPRR107 pKa = 11.84DD108 pKa = 3.93LGPAGPRR115 pKa = 11.84SMSVTLEE122 pKa = 3.52IDD124 pKa = 3.5PPGSVANYY132 pKa = 7.14MVQAAGLSDD141 pKa = 4.52DD142 pKa = 5.59DD143 pKa = 4.18YY144 pKa = 11.78FNVGGSGLAFNPEE157 pKa = 4.24PVNDD161 pKa = 3.98TDD163 pKa = 4.49LLEE166 pKa = 4.27AAEE169 pKa = 5.59DD170 pKa = 3.73ILSLSPIGDD179 pKa = 3.52PEE181 pKa = 4.26MPGSGDD187 pKa = 3.46EE188 pKa = 4.39LIEE191 pKa = 4.66LIEE194 pKa = 4.59TIASQLKK201 pKa = 9.02NWTASAEE208 pKa = 4.25SPMEE212 pKa = 4.14VFVHH216 pKa = 5.67QAATIEE222 pKa = 4.58GIYY225 pKa = 10.76VNGEE229 pKa = 4.16LVTEE233 pKa = 4.26APKK236 pKa = 10.94LSDD239 pKa = 3.16YY240 pKa = 11.31HH241 pKa = 8.58SFDD244 pKa = 4.99DD245 pKa = 5.21EE246 pKa = 4.85EE247 pKa = 5.44EE248 pKa = 5.13GDD250 pKa = 5.05DD251 pKa = 4.89DD252 pKa = 4.87TDD254 pKa = 5.24DD255 pKa = 4.41GPAPNVVVHH264 pKa = 7.07ADD266 pKa = 3.41GTVTIDD272 pKa = 3.24VSVEE276 pKa = 3.89VVAGGNTVVNNAVMQNLWTAATVTAVLGDD305 pKa = 3.7HH306 pKa = 6.69FEE308 pKa = 4.32INAIIQTNAIWDD320 pKa = 3.72TDD322 pKa = 4.34AITSAIEE329 pKa = 3.7SWTNSAGANEE339 pKa = 4.54IFNIATFEE347 pKa = 4.1RR348 pKa = 11.84MDD350 pKa = 3.47SSEE353 pKa = 4.23DD354 pKa = 3.17TGAQPVTGGFPSAWAITEE372 pKa = 3.94IQGDD376 pKa = 3.95LMIANWLEE384 pKa = 3.7QYY386 pKa = 10.76IFVSDD391 pKa = 3.69NDD393 pKa = 3.48IGILSSSGVTTSVIAGDD410 pKa = 3.72NTSTNHH416 pKa = 5.22VSLYY420 pKa = 10.55EE421 pKa = 4.22LGLSYY426 pKa = 11.29DD427 pKa = 4.49LIIVGGSVYY436 pKa = 10.41DD437 pKa = 3.82ANIIHH442 pKa = 6.0QFNLLFDD449 pKa = 4.39NDD451 pKa = 3.59VVGAVSGFEE460 pKa = 4.23TTGEE464 pKa = 4.33GAFSGAGNLLWNQAYY479 pKa = 9.54IYY481 pKa = 10.2NIGGADD487 pKa = 3.72RR488 pKa = 11.84FDD490 pKa = 4.84ALPSAYY496 pKa = 10.22LDD498 pKa = 3.7AANGIAAGRR507 pKa = 11.84NSLPNGVLTDD517 pKa = 3.74PAFGGMEE524 pKa = 4.35GLRR527 pKa = 11.84VLYY530 pKa = 10.36ISGDD534 pKa = 3.74LLNIQYY540 pKa = 9.76IRR542 pKa = 11.84QTTIVGDD549 pKa = 3.95DD550 pKa = 3.77DD551 pKa = 4.94QIALAMNAVNPHH563 pKa = 7.1PDD565 pKa = 3.24ASWSVSTGDD574 pKa = 3.25NTLVNTAAILDD585 pKa = 4.36LDD587 pKa = 4.41SMGKK591 pKa = 8.03TYY593 pKa = 11.0VGGEE597 pKa = 4.09QYY599 pKa = 10.88SQEE602 pKa = 4.08TLIQAEE608 pKa = 5.19LISSDD613 pKa = 3.88PEE615 pKa = 3.84LAARR619 pKa = 11.84DD620 pKa = 3.75PNEE623 pKa = 4.12LVNEE627 pKa = 4.1AVVFLDD633 pKa = 5.08DD634 pKa = 4.84SMLTAEE640 pKa = 4.83TDD642 pKa = 3.68AQGLPAPEE650 pKa = 5.02HH651 pKa = 6.78DD652 pKa = 3.64SHH654 pKa = 6.41YY655 pKa = 11.15QNDD658 pKa = 4.36GLQHH662 pKa = 5.34VLGG665 pKa = 4.52
Molecular weight: 71.22 kDa
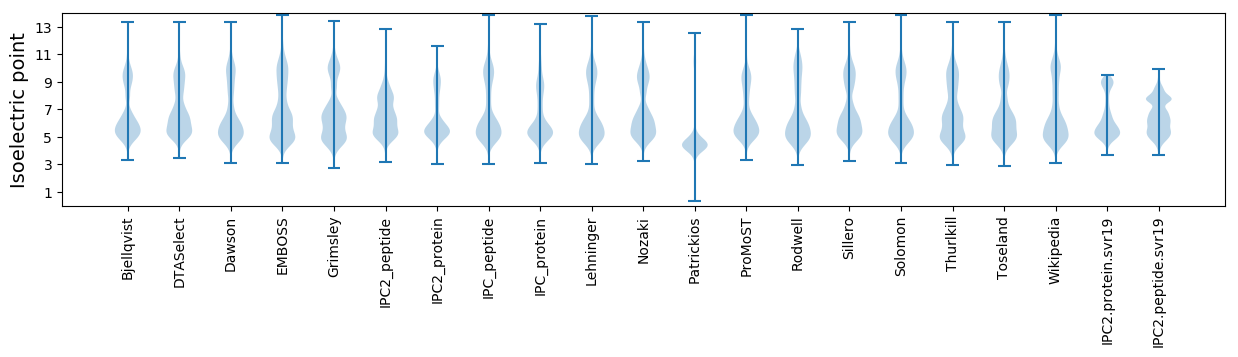
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M3BRG3|A0A0M3BRG3_9RHIZ GntR family transcriptional regulator OS=Rhizobium sp. LC145 OX=1120688 GN=YH62_07460 PE=3 SV=1
MM1 pKa = 7.02MMAMLTRR8 pKa = 11.84MRR10 pKa = 11.84MPTQTQTLMRR20 pKa = 11.84MLTPMQTPMRR30 pKa = 11.84MRR32 pKa = 11.84TLMPMLMPTRR42 pKa = 11.84TQMLMPMPMRR52 pKa = 11.84MPTLMQMLMPMPMRR66 pKa = 11.84MPTLMQMQMLTLMQMLTPTLMRR88 pKa = 11.84MPTQTLTPMLMRR100 pKa = 11.84MLTLMQMRR108 pKa = 11.84MRR110 pKa = 11.84MPTLMRR116 pKa = 11.84MRR118 pKa = 11.84MLTPMRR124 pKa = 11.84MPTPTQMLMRR134 pKa = 11.84MPTQTQTLTPTLTRR148 pKa = 11.84MQTPTQMLMRR158 pKa = 11.84MPTLMPMRR166 pKa = 11.84TQMLMQMLTPTLMRR180 pKa = 11.84MPTQTLTPMLMRR192 pKa = 11.84TLMQMQMLTRR202 pKa = 11.84MQTPMQMLMRR212 pKa = 11.84MPTLMRR218 pKa = 11.84MPMLMPMPMPTRR230 pKa = 11.84TPMRR234 pKa = 11.84MRR236 pKa = 11.84TQMPTQTLMPMPMLMRR252 pKa = 11.84TQMQMLTPMRR262 pKa = 11.84MQTPTLMPMLMLMQMRR278 pKa = 11.84MPTLMPMLTLTPMRR292 pKa = 11.84MPTLTPMRR300 pKa = 11.84MPTLTPMRR308 pKa = 11.84MPTRR312 pKa = 11.84TLTPMRR318 pKa = 11.84TPMQTPMRR326 pKa = 11.84TQMPMPMPTLMQTLTPMRR344 pKa = 11.84MRR346 pKa = 11.84MLTLTPMRR354 pKa = 11.84MPTLTPMRR362 pKa = 11.84MPTRR366 pKa = 11.84TPMRR370 pKa = 11.84TLMRR374 pKa = 11.84MRR376 pKa = 11.84MLLMILLPRR385 pKa = 11.84RR386 pKa = 11.84LSSAPSPARR395 pKa = 11.84EE396 pKa = 4.27QISPPTPHH404 pKa = 6.53SS405 pKa = 3.46
MM1 pKa = 7.02MMAMLTRR8 pKa = 11.84MRR10 pKa = 11.84MPTQTQTLMRR20 pKa = 11.84MLTPMQTPMRR30 pKa = 11.84MRR32 pKa = 11.84TLMPMLMPTRR42 pKa = 11.84TQMLMPMPMRR52 pKa = 11.84MPTLMQMLMPMPMRR66 pKa = 11.84MPTLMQMQMLTLMQMLTPTLMRR88 pKa = 11.84MPTQTLTPMLMRR100 pKa = 11.84MLTLMQMRR108 pKa = 11.84MRR110 pKa = 11.84MPTLMRR116 pKa = 11.84MRR118 pKa = 11.84MLTPMRR124 pKa = 11.84MPTPTQMLMRR134 pKa = 11.84MPTQTQTLTPTLTRR148 pKa = 11.84MQTPTQMLMRR158 pKa = 11.84MPTLMPMRR166 pKa = 11.84TQMLMQMLTPTLMRR180 pKa = 11.84MPTQTLTPMLMRR192 pKa = 11.84TLMQMQMLTRR202 pKa = 11.84MQTPMQMLMRR212 pKa = 11.84MPTLMRR218 pKa = 11.84MPMLMPMPMPTRR230 pKa = 11.84TPMRR234 pKa = 11.84MRR236 pKa = 11.84TQMPTQTLMPMPMLMRR252 pKa = 11.84TQMQMLTPMRR262 pKa = 11.84MQTPTLMPMLMLMQMRR278 pKa = 11.84MPTLMPMLTLTPMRR292 pKa = 11.84MPTLTPMRR300 pKa = 11.84MPTLTPMRR308 pKa = 11.84MPTRR312 pKa = 11.84TLTPMRR318 pKa = 11.84TPMQTPMRR326 pKa = 11.84TQMPMPMPTLMQTLTPMRR344 pKa = 11.84MRR346 pKa = 11.84MLTLTPMRR354 pKa = 11.84MPTLTPMRR362 pKa = 11.84MPTRR366 pKa = 11.84TPMRR370 pKa = 11.84TLMRR374 pKa = 11.84MRR376 pKa = 11.84MLLMILLPRR385 pKa = 11.84RR386 pKa = 11.84LSSAPSPARR395 pKa = 11.84EE396 pKa = 4.27QISPPTPHH404 pKa = 6.53SS405 pKa = 3.46
Molecular weight: 48.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1692521 |
28 |
2835 |
302.1 |
32.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.851 ± 0.044 | 0.747 ± 0.011 |
5.455 ± 0.025 | 6.194 ± 0.035 |
3.972 ± 0.023 | 8.396 ± 0.03 |
1.981 ± 0.016 | 5.739 ± 0.023 |
3.619 ± 0.029 | 9.983 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.642 ± 0.017 | 2.754 ± 0.018 |
4.985 ± 0.023 | 3.028 ± 0.019 |
6.856 ± 0.037 | 5.672 ± 0.019 |
5.207 ± 0.024 | 7.301 ± 0.028 |
1.303 ± 0.013 | 2.313 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
