
Streptococcus satellite phage Javan379
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
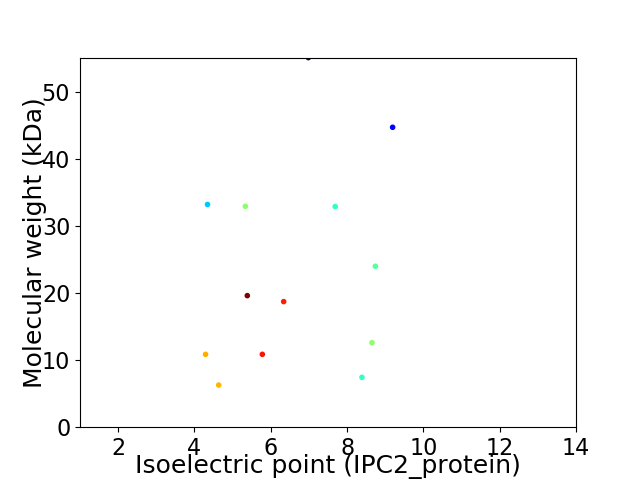
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZRD6|A0A4D5ZRD6_9VIRU HTH cro/C1-type domain-containing protein OS=Streptococcus satellite phage Javan379 OX=2558669 GN=JavanS379_0011 PE=4 SV=1
MM1 pKa = 7.4IGNYY5 pKa = 9.7LKK7 pKa = 10.7NLRR10 pKa = 11.84KK11 pKa = 9.32RR12 pKa = 11.84QNIKK16 pKa = 10.23VVEE19 pKa = 4.06LAKK22 pKa = 9.78TVQVSQPYY30 pKa = 9.63ISNIEE35 pKa = 3.86NEE37 pKa = 3.92KK38 pKa = 10.56RR39 pKa = 11.84YY40 pKa = 6.91PTKK43 pKa = 10.88DD44 pKa = 3.2LFFKK48 pKa = 10.61IIISIAEE55 pKa = 4.31LSPLNYY61 pKa = 9.63EE62 pKa = 4.45VYY64 pKa = 10.76CDD66 pKa = 4.14LEE68 pKa = 4.06LSKK71 pKa = 10.6EE72 pKa = 4.12QKK74 pKa = 10.04KK75 pKa = 10.63DD76 pKa = 2.89IYY78 pKa = 10.68IPDD81 pKa = 3.68VLPEE85 pKa = 4.04FWEE88 pKa = 5.03KK89 pKa = 10.89YY90 pKa = 10.2SSTILKK96 pKa = 10.08NINEE100 pKa = 4.21YY101 pKa = 11.0LDD103 pKa = 5.07DD104 pKa = 4.39EE105 pKa = 5.17DD106 pKa = 5.71EE107 pKa = 4.22ITSFEE112 pKa = 4.11DD113 pKa = 3.45FKK115 pKa = 11.5DD116 pKa = 4.37YY117 pKa = 11.04VSSWNIEE124 pKa = 3.83DD125 pKa = 3.46MAIFPEE131 pKa = 4.53FTDD134 pKa = 4.53FLDD137 pKa = 3.87SKK139 pKa = 10.99YY140 pKa = 11.12GQDD143 pKa = 4.44DD144 pKa = 3.55YY145 pKa = 12.2GEE147 pKa = 4.14FLDD150 pKa = 3.9YY151 pKa = 10.92KK152 pKa = 10.61DD153 pKa = 3.72YY154 pKa = 11.55SSYY157 pKa = 11.14QDD159 pKa = 3.26PEE161 pKa = 4.41YY162 pKa = 10.51IKK164 pKa = 10.92SIVTDD169 pKa = 2.87YY170 pKa = 10.46WYY172 pKa = 10.86QDD174 pKa = 3.41FLTEE178 pKa = 4.07FLDD181 pKa = 3.88YY182 pKa = 11.16FEE184 pKa = 4.73NTSTIDD190 pKa = 4.09GIPNPEE196 pKa = 3.93YY197 pKa = 10.68TEE199 pKa = 4.14IMLEE203 pKa = 3.97TLAPQEE209 pKa = 4.08LEE211 pKa = 3.51IYY213 pKa = 7.31TTIKK217 pKa = 10.19EE218 pKa = 4.13LQHH221 pKa = 6.45RR222 pKa = 11.84KK223 pKa = 10.32GIFDD227 pKa = 4.07RR228 pKa = 11.84YY229 pKa = 8.25QFKK232 pKa = 9.14NTDD235 pKa = 3.52KK236 pKa = 11.64NNLTDD241 pKa = 5.18LSIIDD246 pKa = 4.49DD247 pKa = 3.86SEE249 pKa = 5.71AIYY252 pKa = 11.05KK253 pKa = 8.4LTLDD257 pKa = 4.16GKK259 pKa = 9.27HH260 pKa = 6.25LSSADD265 pKa = 3.27IIALQTLINGIRR277 pKa = 11.84YY278 pKa = 8.9KK279 pKa = 10.69RR280 pKa = 3.43
MM1 pKa = 7.4IGNYY5 pKa = 9.7LKK7 pKa = 10.7NLRR10 pKa = 11.84KK11 pKa = 9.32RR12 pKa = 11.84QNIKK16 pKa = 10.23VVEE19 pKa = 4.06LAKK22 pKa = 9.78TVQVSQPYY30 pKa = 9.63ISNIEE35 pKa = 3.86NEE37 pKa = 3.92KK38 pKa = 10.56RR39 pKa = 11.84YY40 pKa = 6.91PTKK43 pKa = 10.88DD44 pKa = 3.2LFFKK48 pKa = 10.61IIISIAEE55 pKa = 4.31LSPLNYY61 pKa = 9.63EE62 pKa = 4.45VYY64 pKa = 10.76CDD66 pKa = 4.14LEE68 pKa = 4.06LSKK71 pKa = 10.6EE72 pKa = 4.12QKK74 pKa = 10.04KK75 pKa = 10.63DD76 pKa = 2.89IYY78 pKa = 10.68IPDD81 pKa = 3.68VLPEE85 pKa = 4.04FWEE88 pKa = 5.03KK89 pKa = 10.89YY90 pKa = 10.2SSTILKK96 pKa = 10.08NINEE100 pKa = 4.21YY101 pKa = 11.0LDD103 pKa = 5.07DD104 pKa = 4.39EE105 pKa = 5.17DD106 pKa = 5.71EE107 pKa = 4.22ITSFEE112 pKa = 4.11DD113 pKa = 3.45FKK115 pKa = 11.5DD116 pKa = 4.37YY117 pKa = 11.04VSSWNIEE124 pKa = 3.83DD125 pKa = 3.46MAIFPEE131 pKa = 4.53FTDD134 pKa = 4.53FLDD137 pKa = 3.87SKK139 pKa = 10.99YY140 pKa = 11.12GQDD143 pKa = 4.44DD144 pKa = 3.55YY145 pKa = 12.2GEE147 pKa = 4.14FLDD150 pKa = 3.9YY151 pKa = 10.92KK152 pKa = 10.61DD153 pKa = 3.72YY154 pKa = 11.55SSYY157 pKa = 11.14QDD159 pKa = 3.26PEE161 pKa = 4.41YY162 pKa = 10.51IKK164 pKa = 10.92SIVTDD169 pKa = 2.87YY170 pKa = 10.46WYY172 pKa = 10.86QDD174 pKa = 3.41FLTEE178 pKa = 4.07FLDD181 pKa = 3.88YY182 pKa = 11.16FEE184 pKa = 4.73NTSTIDD190 pKa = 4.09GIPNPEE196 pKa = 3.93YY197 pKa = 10.68TEE199 pKa = 4.14IMLEE203 pKa = 3.97TLAPQEE209 pKa = 4.08LEE211 pKa = 3.51IYY213 pKa = 7.31TTIKK217 pKa = 10.19EE218 pKa = 4.13LQHH221 pKa = 6.45RR222 pKa = 11.84KK223 pKa = 10.32GIFDD227 pKa = 4.07RR228 pKa = 11.84YY229 pKa = 8.25QFKK232 pKa = 9.14NTDD235 pKa = 3.52KK236 pKa = 11.64NNLTDD241 pKa = 5.18LSIIDD246 pKa = 4.49DD247 pKa = 3.86SEE249 pKa = 5.71AIYY252 pKa = 11.05KK253 pKa = 8.4LTLDD257 pKa = 4.16GKK259 pKa = 9.27HH260 pKa = 6.25LSSADD265 pKa = 3.27IIALQTLINGIRR277 pKa = 11.84YY278 pKa = 8.9KK279 pKa = 10.69RR280 pKa = 3.43
Molecular weight: 33.27 kDa
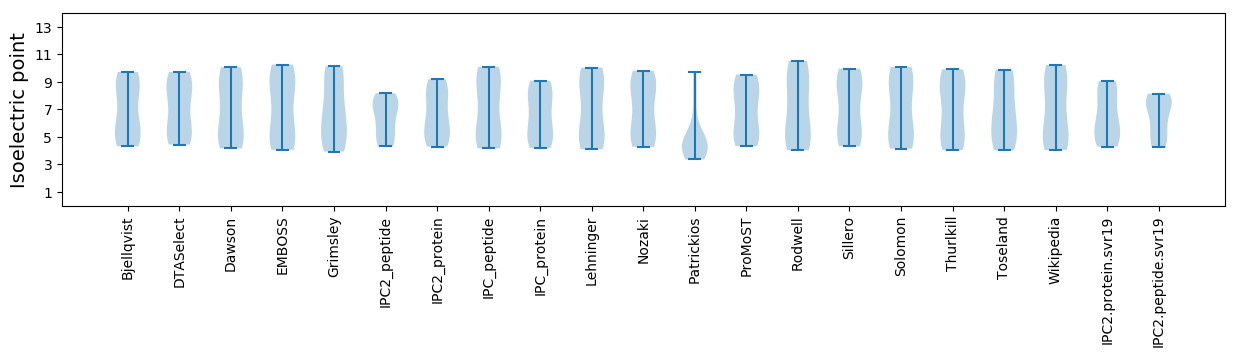
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZLV8|A0A4D5ZLV8_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan379 OX=2558669 GN=JavanS379_0002 PE=4 SV=1
MM1 pKa = 7.99KK2 pKa = 9.16ITQHH6 pKa = 5.97IKK8 pKa = 8.98QDD10 pKa = 3.16GSIVYY15 pKa = 10.06RR16 pKa = 11.84SSIYY20 pKa = 10.7LGIDD24 pKa = 3.12QITGKK29 pKa = 10.06QVTTKK34 pKa = 9.23ITGRR38 pKa = 11.84TKK40 pKa = 10.86KK41 pKa = 9.71EE42 pKa = 4.06VKK44 pKa = 10.06QKK46 pKa = 11.0AQDD49 pKa = 3.3AVIDD53 pKa = 4.07FKK55 pKa = 11.51INGSTRR61 pKa = 11.84FQASTISTYY70 pKa = 11.16EE71 pKa = 3.9EE72 pKa = 4.49LASLWWDD79 pKa = 3.4NHH81 pKa = 3.94KK82 pKa = 9.36HH83 pKa = 4.09TVKK86 pKa = 10.56PNSQDD91 pKa = 2.89ATKK94 pKa = 10.73RR95 pKa = 11.84LIDD98 pKa = 3.61NHH100 pKa = 5.95VLPLFGAYY108 pKa = 9.56KK109 pKa = 10.2LDD111 pKa = 3.65KK112 pKa = 10.12LTTPLIQNIVNKK124 pKa = 10.34LADD127 pKa = 3.6KK128 pKa = 9.52TNKK131 pKa = 10.12GEE133 pKa = 4.13AGAYY137 pKa = 8.89LHH139 pKa = 6.56YY140 pKa = 11.05DD141 pKa = 4.09KK142 pKa = 11.05IHH144 pKa = 6.58ALNKK148 pKa = 10.18RR149 pKa = 11.84ILQYY153 pKa = 10.84GVVMQAIPFNPARR166 pKa = 11.84EE167 pKa = 4.24VILPRR172 pKa = 11.84NTKK175 pKa = 9.05KK176 pKa = 10.8ANRR179 pKa = 11.84QKK181 pKa = 10.89VKK183 pKa = 10.57HH184 pKa = 5.76FNNLEE189 pKa = 4.01LKK191 pKa = 10.65RR192 pKa = 11.84FLDD195 pKa = 4.03YY196 pKa = 11.21LDD198 pKa = 4.46SLNTSKK204 pKa = 10.76YY205 pKa = 8.48RR206 pKa = 11.84YY207 pKa = 8.75FYY209 pKa = 10.96EE210 pKa = 3.79VTLYY214 pKa = 11.08KK215 pKa = 10.63FILATGCRR223 pKa = 11.84INEE226 pKa = 3.95VLALEE231 pKa = 4.43WSDD234 pKa = 3.56IDD236 pKa = 4.22LDD238 pKa = 4.0SAVVHH243 pKa = 5.01VTKK246 pKa = 10.16TLNYY250 pKa = 8.35KK251 pKa = 9.4QEE253 pKa = 4.1INSPKK258 pKa = 10.23SKK260 pKa = 10.48SSYY263 pKa = 10.28RR264 pKa = 11.84DD265 pKa = 2.84IDD267 pKa = 3.35IDD269 pKa = 3.81NQTVTMLKK277 pKa = 9.66QYY279 pKa = 10.52KK280 pKa = 9.62RR281 pKa = 11.84RR282 pKa = 11.84QTQEE286 pKa = 3.07AWKK289 pKa = 10.2LGQTEE294 pKa = 4.85SIVFSDD300 pKa = 5.45FINGYY305 pKa = 10.27ANNRR309 pKa = 11.84SLFTRR314 pKa = 11.84LTTHH318 pKa = 6.89FKK320 pKa = 10.02RR321 pKa = 11.84AKK323 pKa = 8.34VPNIGFHH330 pKa = 5.98GFRR333 pKa = 11.84HH334 pKa = 4.87THH336 pKa = 6.58ASLLLNSGIPYY347 pKa = 10.0KK348 pKa = 10.34EE349 pKa = 3.8LQHH352 pKa = 6.73RR353 pKa = 11.84LGHH356 pKa = 5.68STLSMTMDD364 pKa = 3.58IYY366 pKa = 11.68SHH368 pKa = 6.98LSKK371 pKa = 11.0EE372 pKa = 4.25NAKK375 pKa = 10.37KK376 pKa = 10.38AVSFYY381 pKa = 10.59EE382 pKa = 3.87MAMEE386 pKa = 5.16SII388 pKa = 4.37
MM1 pKa = 7.99KK2 pKa = 9.16ITQHH6 pKa = 5.97IKK8 pKa = 8.98QDD10 pKa = 3.16GSIVYY15 pKa = 10.06RR16 pKa = 11.84SSIYY20 pKa = 10.7LGIDD24 pKa = 3.12QITGKK29 pKa = 10.06QVTTKK34 pKa = 9.23ITGRR38 pKa = 11.84TKK40 pKa = 10.86KK41 pKa = 9.71EE42 pKa = 4.06VKK44 pKa = 10.06QKK46 pKa = 11.0AQDD49 pKa = 3.3AVIDD53 pKa = 4.07FKK55 pKa = 11.51INGSTRR61 pKa = 11.84FQASTISTYY70 pKa = 11.16EE71 pKa = 3.9EE72 pKa = 4.49LASLWWDD79 pKa = 3.4NHH81 pKa = 3.94KK82 pKa = 9.36HH83 pKa = 4.09TVKK86 pKa = 10.56PNSQDD91 pKa = 2.89ATKK94 pKa = 10.73RR95 pKa = 11.84LIDD98 pKa = 3.61NHH100 pKa = 5.95VLPLFGAYY108 pKa = 9.56KK109 pKa = 10.2LDD111 pKa = 3.65KK112 pKa = 10.12LTTPLIQNIVNKK124 pKa = 10.34LADD127 pKa = 3.6KK128 pKa = 9.52TNKK131 pKa = 10.12GEE133 pKa = 4.13AGAYY137 pKa = 8.89LHH139 pKa = 6.56YY140 pKa = 11.05DD141 pKa = 4.09KK142 pKa = 11.05IHH144 pKa = 6.58ALNKK148 pKa = 10.18RR149 pKa = 11.84ILQYY153 pKa = 10.84GVVMQAIPFNPARR166 pKa = 11.84EE167 pKa = 4.24VILPRR172 pKa = 11.84NTKK175 pKa = 9.05KK176 pKa = 10.8ANRR179 pKa = 11.84QKK181 pKa = 10.89VKK183 pKa = 10.57HH184 pKa = 5.76FNNLEE189 pKa = 4.01LKK191 pKa = 10.65RR192 pKa = 11.84FLDD195 pKa = 4.03YY196 pKa = 11.21LDD198 pKa = 4.46SLNTSKK204 pKa = 10.76YY205 pKa = 8.48RR206 pKa = 11.84YY207 pKa = 8.75FYY209 pKa = 10.96EE210 pKa = 3.79VTLYY214 pKa = 11.08KK215 pKa = 10.63FILATGCRR223 pKa = 11.84INEE226 pKa = 3.95VLALEE231 pKa = 4.43WSDD234 pKa = 3.56IDD236 pKa = 4.22LDD238 pKa = 4.0SAVVHH243 pKa = 5.01VTKK246 pKa = 10.16TLNYY250 pKa = 8.35KK251 pKa = 9.4QEE253 pKa = 4.1INSPKK258 pKa = 10.23SKK260 pKa = 10.48SSYY263 pKa = 10.28RR264 pKa = 11.84DD265 pKa = 2.84IDD267 pKa = 3.35IDD269 pKa = 3.81NQTVTMLKK277 pKa = 9.66QYY279 pKa = 10.52KK280 pKa = 9.62RR281 pKa = 11.84RR282 pKa = 11.84QTQEE286 pKa = 3.07AWKK289 pKa = 10.2LGQTEE294 pKa = 4.85SIVFSDD300 pKa = 5.45FINGYY305 pKa = 10.27ANNRR309 pKa = 11.84SLFTRR314 pKa = 11.84LTTHH318 pKa = 6.89FKK320 pKa = 10.02RR321 pKa = 11.84AKK323 pKa = 8.34VPNIGFHH330 pKa = 5.98GFRR333 pKa = 11.84HH334 pKa = 4.87THH336 pKa = 6.58ASLLLNSGIPYY347 pKa = 10.0KK348 pKa = 10.34EE349 pKa = 3.8LQHH352 pKa = 6.73RR353 pKa = 11.84LGHH356 pKa = 5.68STLSMTMDD364 pKa = 3.58IYY366 pKa = 11.68SHH368 pKa = 6.98LSKK371 pKa = 11.0EE372 pKa = 4.25NAKK375 pKa = 10.37KK376 pKa = 10.38AVSFYY381 pKa = 10.59EE382 pKa = 3.87MAMEE386 pKa = 5.16SII388 pKa = 4.37
Molecular weight: 44.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2678 |
55 |
479 |
206.0 |
23.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.452 ± 0.447 | 0.523 ± 0.127 |
5.601 ± 0.562 | 8.364 ± 0.713 |
4.294 ± 0.278 | 4.668 ± 0.365 |
1.755 ± 0.356 | 6.945 ± 0.458 |
8.701 ± 0.435 | 9.821 ± 0.545 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.166 ± 0.266 | 6.273 ± 0.54 |
3.174 ± 0.61 | 4.444 ± 0.418 |
4.742 ± 0.337 | 5.639 ± 0.441 |
6.535 ± 0.325 | 5.078 ± 0.493 |
0.859 ± 0.156 | 4.966 ± 0.538 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
